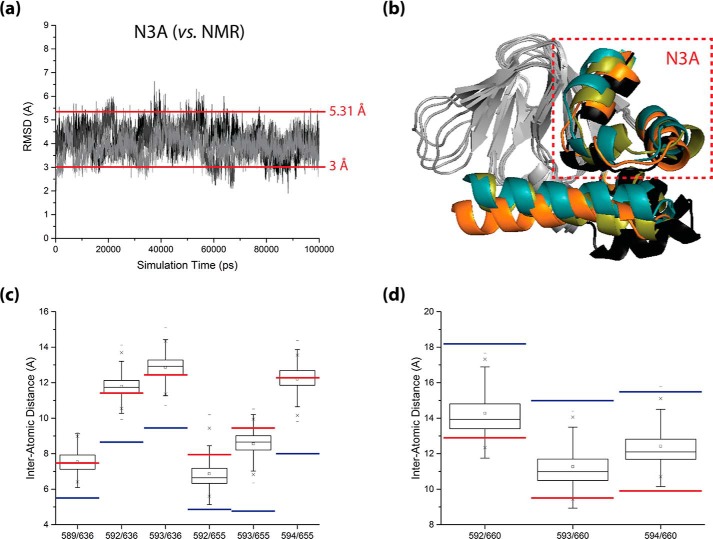
FIGURE 5.
Sampling of inactive-state-like CBD topologies within the apo-monomer HCN4 IR simulations. a, backbone root-mean-square deviation trajectories of the N3A (i.e. αE′-αA region) from its configuration in the average apo-state NMR structure of the HCN4 CBD (i.e. the average structure derived from RCSB PDB entry 2MNG), as observed in each of the three replicate apo-monomer HCN4 IR simulations (black, dark gray, and light gray plots, respectively). All root-mean-square deviations were computed with structure overlay at the β-core. The threshold r.m.s.d. value used for selecting inactive-state-like structures for further assessment (i.e. 3 Å) and the r.m.s.d. between the 3OTF and average 2MNG structures (i.e. 5.31 Å), are indicated by horizontal lines. b, ribbon structure illustration of the selected inactive-state-like structures exhibiting the smallest (orange ribbon), median (olive green ribbon), and largest (teal ribbon) N3A root-mean-square deviations from the average 2MNG structure, overlaid at their β-cores onto the average 2MNG structure (black ribbon). For clarity, the β-cores of all structures are shown in gray, and the αA′–αD′ helices of all structures are omitted. c and d, boxplots of inter-α-carbon distances for selected pairs of residues that bridge the interface between the N3A and β-core, as observed for the selected ensemble of inactive-state-like structures. Residue pairs that are farther apart in the average 2MNG structure (c) and residue pairs that are closer together in the average 2MNG structure (d) were considered. All boxplots were constructed using Origin 9.1 (OriginLab Corp.), and for reference, the corresponding distance values observed in the 3OTF structure (blue lines) and in the average 2MNG structure (red lines) are indicated for all plots.