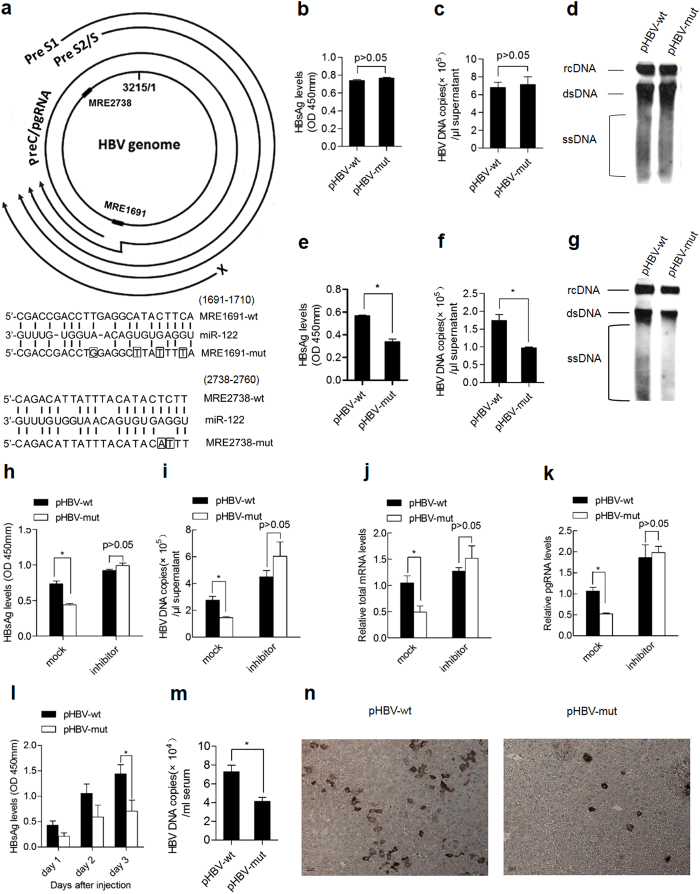
Figure 1. miR-122 response elements facilitate viral replication in hepatocytes with high miR-122 abundance.
(a) (Top panel) Schematic representation of putative miR-122 response elements in all four HBV mRNAs. ■ represents the miR-122 response elements. (Lower panel) Diagram of predicted miR-122 binding sequence located in the HBV genome. Perfect matches are indicated by a line. Mutations (the frame) were made in the seed region of the miR-122 binding sites. (b–g) HepG2 (b–d) and Huh7 (e–g) cells were transfected with pHBV-wt or pHBV-mut plasmid .The levels of HBsAg in the supernatant were detected by ELISA 48 h after transfection (b,e) and DNA copies were quantified by real-time PCR (c,f) HBV DNA levels in cells were measured by Southern blotting (d,g) (h–k) Huh7 cells were co-transfected with pHBV-wt or pHBV-mut plasmid and 122 inhibitor or a randomized oligonucleotide (mock). The secretion of HBsAg was measured by ELISA (h) HBV DNA copies (i), total mRNA (j) and pgRNA levels (k) were quantified by real-time PCR. (l–n) Female BALB/c mice were injected with 15 μg pHBV-wt or pHBV-mut plasmid via the tail vein using the hydrodynamic method (five mice per group). Serum HBsAg was measured at the indicated time point (l) HBV DNA copies in serum were determined 3 d after hydrodynamic injection (m) HBcAg expression in mice livers was analyzed by immunohistochemical staining 3 d after hydrodynamic injection (n) Data shown are the means ± SDs of five mice. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01.