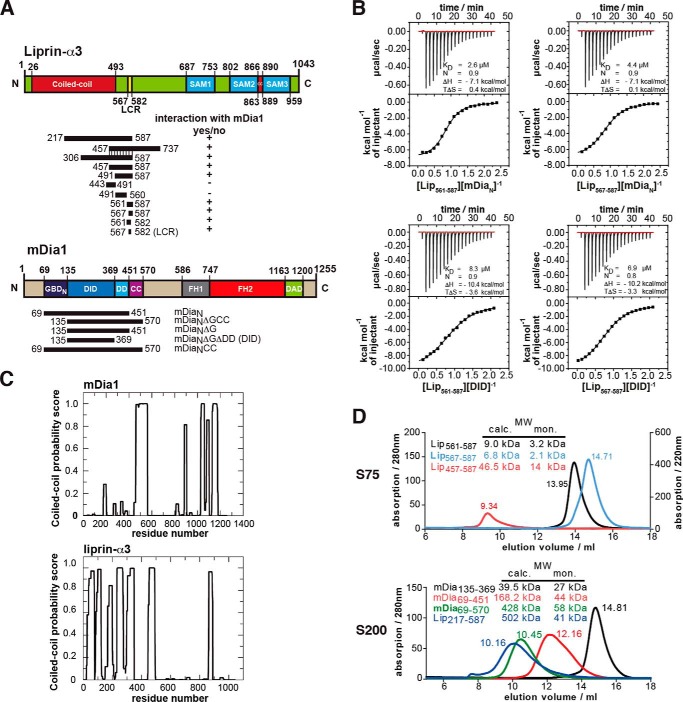
FIGURE 1.
Definition of the binding sites in liprin-α3 and mDia1. A, domain organization of liprin-α3 and mDia1 and the constructs used in this study. +, interactions of liprin-α3 fragments with mDia1; −, fragments not interacting. B, thermodynamic characterization of the shortest liprin-α3 and mDia1 fragments needed for full interaction using ITC. Lip(567–587) and DID bind with 6.9 μm, showing that those fragments contain all residues essential for the interaction. C, coiled-coil prediction of mDia1 and mouse liprin-α3 using COILS version 2.1. For mDia1, the prediction shows a high score for aa 460–562 forming a coiled-coil. For liprin-α3, extended coiled-coil regions were predicted in the N terminus covering residues aa 26–493 and a second patch in the C terminus from aa 863 to 889. D, analytical size exclusion chromatography of mDia1 and liprin-α3 fragments as indicated. Shown is the molecular size calculated by calibration (MW calc.) and the molecular weights for the respective monomers (MW mon.). For all proteins, the absorption at 280 nm is shown except for Lip(567–587) (for which A220 nm is shown). Aprotinin (6.5 kDa), ribonuclease A (13.7 kDa), carbonic anhydrase (29.0 kDa), ovalbumin (43.0 kDa), and conalbumin (75.0 kDa) were used for calibration of the S75 10/300, and for S200 10/300, additionally aldolase (158 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa) were used.