FIGURE 1.
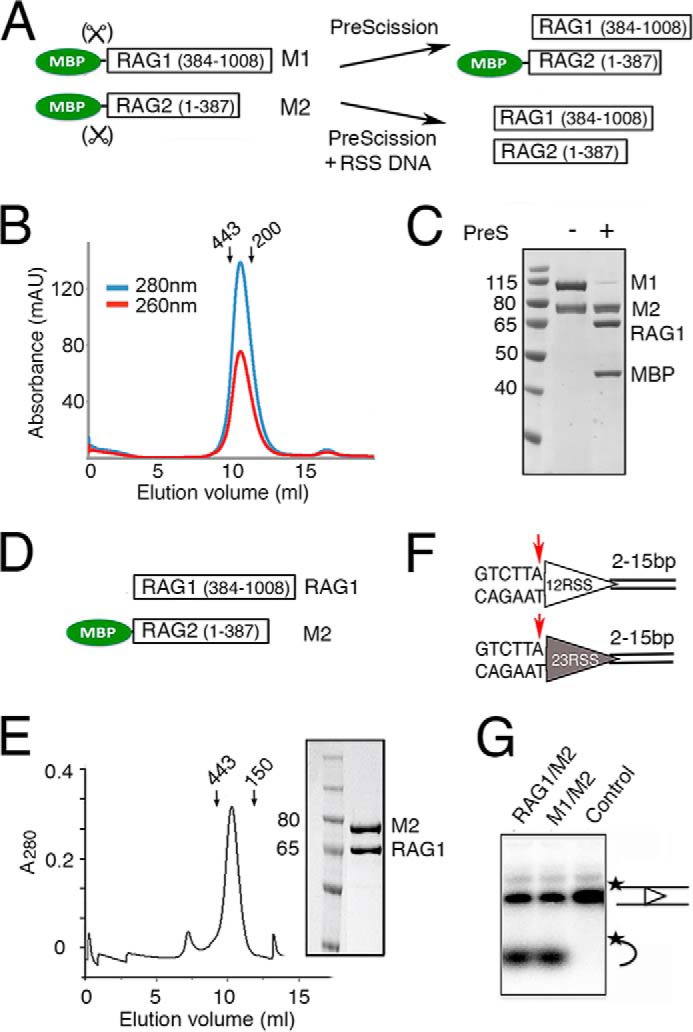
Purification and activity of RAG1 and RAG2 proteins. A, diagram of RAG1 and RAG2 constructs fused to a cleavable MBP tag (M1 and M2). The MBP tag can be efficiently removed from RAG1, but its removal from RAG2 requires binding of 12/23RSS DNAs. B, Superdex-200 elution profile of the tetrameric MBP-tagged RAG1/2 (calculated molecular mass of 406 kDa), which was co-expressed in HEK293GNTI cells and purified by amylose affinity chromatography. Arrows indicate the elution points of molecular mass markers (443 and 200 kDa). C, SDS protein gel of the size exclusion peak showing 1:1 complex of M1 and M2. MBP can be removed from M1 but not M2 by PreScission (PreS). D, diagram of the tag-free RAG1 and non-cleavable MBP-tagged RAG2 (M2) constructs used to produce RAG1/M2 for biochemical studies. E, Superdex-200 elution profile of RAG1/M2 (calculated molecular mass of 312 kDa) and the Coomassie Blue-stained SDS gel of the co-eluted RAG1/M2 protein. F, diagram of the 12RSS (open triangle) and 23RSS (filled triangle) used in paired complex formation and signal-end complex preparation for crystallization. Conserved elements are designated as triangles, and the red arrow is the RAG1/2 cleavage site. The coding flank sequence is shown, and the extensions bordering the other DNA ends were varied in crystallization trials. G, coupled cleavage of 12/23RSS in Mg2+. The 12RSS top strand was 5′-radiolabeled, allowing the detection of hairpin product by a TBE-urea gel. The results of 12/23RSS paired cleavage by RAG1/M2 and M1/M2 were compared. The control lane is DNA alone. The 32P labels on DNA strands are marked with asterisks.
