FIG 1.
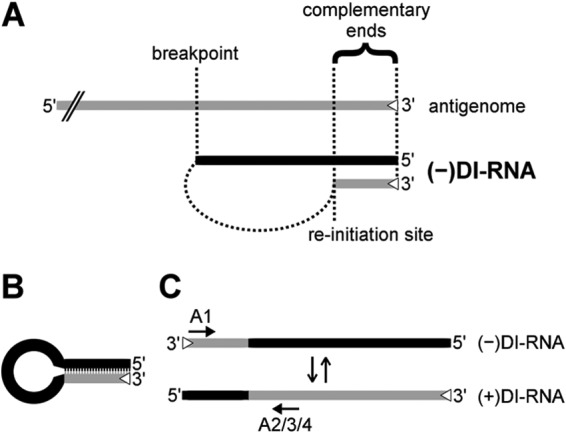
Schematic drawings of 5′ copy-back DI-RNA generation, predicted secondary structure, and replication. (A) 5′ copy-back DI-RNAs are hybrids of truncated genome (black) and antigenome (gray) sequences. During replication, viral polymerase initiates RNA synthesis at the 3′ antigenome promoter (trailer; open triangle) and prematurely terminates synthesis at the breakpoint. The polymerase then reinitiates synthesis on the newly generated strand and finishes replication by recopying the antigenomic 3′ end. The two ends of the DI-RNA are therefore perfectly complementary to each other. (B) Panhandle structure that may be formed through hybridization of the complementary ends of a copy-back DI-RNA in the absence of sufficient N protein. (C) 5′ copy-back DI-RNA replication. The genome DI-RNA [(−)DI-RNA] (top) contains the antigenome promoter in the copy-back sequence (open triangle). Therefore, it can be replicated by the viral polymerase into a complementary antigenome DI-RNA [(+)DI-RNA], which now contains the antigenome promoter at the opposite end. The polarity switch within one DI-RNA molecule allows specific amplification using combinations of primers (primer A1 with either primer A2, A3, or A4; arrows) that would bind in the same direction on a regular antigenome molecule.
