ABSTRACT
Autophagic flux involves formation of autophagosomes and their degradation by lysosomes. Autophagy can either promote or restrict viral replication. In the case of Dengue virus (DENV), several studies report that autophagy supports the viral replication cycle, and describe an increase of autophagic vesicles (AVs) following infection. However, it is unknown how autophagic flux is altered to result in increased AVs. To address this question and gain insight into the role of autophagy during DENV infection, we established an unbiased, image-based flow cytometry approach to quantify autophagic flux under normal growth conditions and in response to activation by nutrient deprivation or the mTOR inhibitor Torin1. We found that DENV induced an initial activation of autophagic flux, followed by inhibition of general and specific autophagy. Early after infection, basal and activated autophagic flux was enhanced. However, during established replication, basal and Torin1-activated autophagic flux was blocked, while autophagic flux activated by nutrient deprivation was reduced, indicating a block to AV formation and reduced AV degradation capacity. During late infection AV levels increased as a result of inefficient fusion of autophagosomes with lysosomes. In addition, endolysosomal trafficking was suppressed, while lysosomal activities were increased. We further determined that DENV infection progressively reduced levels of the autophagy receptor SQSTM1/p62 via proteasomal degradation. Importantly, stable overexpression of p62 significantly suppressed DENV replication, suggesting a novel role for p62 as a viral restriction factor. Overall, our findings indicate that in the course of DENV infection, autophagy shifts from a supporting to an antiviral role, which is countered by DENV.
IMPORTANCE Autophagic flux is a dynamic process starting with the formation of autophagosomes and ending with their degradation after fusion with lysosomes. Autophagy impacts the replication cycle of many viruses. However, thus far the dynamics of autophagy in case of Dengue virus (DENV) infections has not been systematically quantified. Therefore, we used high-content, imaging-based flow cytometry to quantify autophagic flux and endolysosomal trafficking in response to DENV infection. We report that DENV induced an initial activation of autophagic flux, followed by inhibition of general and specific autophagy. Further, lysosomal activity was increased, but endolysosomal trafficking was suppressed confirming the block of autophagic flux. Importantly, we provide evidence that p62, an autophagy receptor, restrict DENV replication and was specifically depleted in DENV-infected cells via increased proteasomal degradation. These results suggest that during DENV infection autophagy shifts from a proviral to an antiviral cellular process, which is counteracted by the virus.
INTRODUCTION
Dengue virus (DENV) is a member of the family Flaviviridae and is responsible for one of the most common infections transmitted to humans by mosquitoes. DENV is a positive-strand enveloped RNA virus, which enters the cell via clathrin-dependent endocytosis (1). RNA translation, replication and virus particle assembly occur at the endoplasmic reticulum (ER) and ER-derived membranes that are induced by the virus in infected cells (2). Owing to their morphologies, these rearranged membrane structures have been designated convoluted membranes and vesicle packets (3). Recent studies have demonstrated that DENV replication requires autophagy (4–8), a process that targets proteins and/or organelles to lysosomes for degradation. Autophagy involves formation of the double-membrane autophagosome, which sequesters target cytosolic content and then fuses with the lysosome to form an autolysosome where sequestered components are degraded (9). Autophagy is induced upon activation of the class III phosphatidylinositol 3-kinase (PI3K)-Beclin1 complex, which signals formation of the isolation membrane and recruitment of cytosolic autophagy factors, which build the autophagosome. During the maturation process, the cytosolic microtubule-associated protein light chain 3 (LC3-I) is conjugated to phosphatidylethanolamine, and the lipidated form of LC3 (LC3-II) is attached to the autophagosome membrane. The membrane-associated LC3-II provides docking sites for receptors, such as SQSTM1/p62 or NDP52/Calcoco2, that target ubiquitinylated cargo to the autophagosome during selective autophagy (10, 11). This process is also important for the maturation of the autophagosome (12). After closure of the autophagosome, the vesicle fuses with endosomes/lysosomes to form an amphisome. At this stage, lysosomal hydrolases degrade the earlier loaded content.
Autophagy is a crucial component of immunity-linked pathway activities, including NF-κB signaling, the antioxidant response (13), and the generation of viral peptides for presentation via major histocompatibility complex class I (MHC-I) and MHC-II (14, 15). Importantly, viruses can manipulate the autophagic pathway in order to promote different aspects of the viral replication cycle, ranging from virus entry up to egress (16). It is well established that positive-strand RNA viruses utilize autophagy to promote viral RNA translation, RNA replication, and virus particle production (17–24). Several lines of evidence suggest a proviral role of autophagy during DENV infection. Autophagy-deficient fibroblasts have 3-fold decrease in DENV production (4), inhibition of autophagy using 3-methyladenine (3MA) or gene knockdown of autophagy mediators (4–8) limits DENV replication, and the autophagic degradation of lipid droplets is required for DENV replication (7).
Overall, these findings implicate that autophagy supports DENV replication, without affecting RNA translation and virus assembly (7). However, the impact of DENV on autophagic flux, i.e., the dynamics of coupled formation and degradation processes, has not been sufficiently addressed, and it is therefore unclear whether DENV might co-opt only some specific autophagy factors and thus interfere with the whole process. Autophagic flux is determined by the capacity of the cell to degrade forming autophagosomes, which occurs within minutes (25), and AV content increases upon disruption to lysosomal activity. It is commonly reported that DENV infection increases AV number as detected by high-resolution microscopy (6, 7, 24, 26, 27) and Western blot analysis of lipidated LC3-II levels (5, 6, 28, 29). Notably, in autophagosomes colocalizing with lysosomes, pronounced pH-dependent quenching of green fluorescent protein (GFP)-LC3 fluorescence (30) was not observed (7), suggesting that DENV may reduce lysosomal acidification and so disrupt lysosomal function.
Thus, an outstanding question is whether DENV infection increases or suppresses autophagic flux, resulting in an imbalance between formation and degradation rates, and consequently results in the observed increase in AV levels. To test this hypothesis, we developed an unbiased, high-content approach to quantify autophagic flux in cell populations using a high-resolution, image-based flow cytometer. In addition, we studied the effect of DENV on the autophagy receptor SQSTM1/p62 and its role in the viral replication cycle.
MATERIALS AND METHODS
Antibodies.
The polyclonal antibody against DENV NS4B and NS3 was produced as described previously (2, 31). For cellular markers, primary antibodies used were of the following specificities: rabbit polyclonal LC3B and rabbit polyclonal β-actin (Cell Signaling, USA), mouse monoclonal GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Santa Cruz, USA), mouse monoclonal Lamp2 (BD Bioscience, Heidelberg, Germany), mouse monoclonal p62 (BD Bioscience, Heidelberg, Germany), rabbit monoclonal ubiquitin (Cell Signaling), and polyclonal NDP52 (Cell Signaling). Secondary goat antibodies used for indirect immunofluorescence microscopy were conjugated to Alexa Fluor 488 (Molecular Probes, Karlsruhe, Germany) or Cy5 (Jackson ImmunoResearch Laboratories, USA). For all Western blot assays, secondary rabbit or mouse antibodies coupled to horseradish peroxidase (Sigma, Deisenhofen, Germany) were used.
Cell culture.
Huh7 cells (32) and subclones thereof were maintained in Dulbecco modified Eagle medium (DMEM; Invitrogen, Karlsruhe, Germany), supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal calf serum (DMEM cplt). Huh7-GFP-LC3 or Huh7-mCherry-LC3 cells were generated by transduction with lentiviral vectors encoding GFP-LC3 or mCherry-LC3, as previously described (33). Transduced cells were selected and maintained with 5 μg of puromycin (Invitrogen)/ml. At 24 h before the experiments started, puromycin was removed.
Plasmid constructs.
Standard molecular biology techniques were used for cloning, and the nucleotide sequences of all used plasmids were verified. pWPI-Puro-GFP-LC3 and pWPI-Puro-mCherry-LC3 were generated by insertion of a NheI-XbaI fragment, isolated from either mCherry-LC3 or pEGFP-LC3, into the SpeI-linearized vector pWPI-Blr (33). The constructs were propagated in the dam-negative Escherichia coli strain GM2163 to allow restriction digestion of the purified plasmid DNA with XbaI. For pWPI-p62, Gateway technology was used.
Viruses.
Cells were infected with the DENV 2 isolate New Guinea C (kindly provided by Progen Biotechnik GmbH, Heidelberg, Germany). For all infection experiments, virus was diluted in DMEM containing 10% fetal calf serum, and the cells were incubated at 37°C and 5% CO2 with gentle rocking. Two hours later, the inoculum was replaced by DMEM cplt, and the cells were incubated as specified in Results.
Determination of virus titers by plaque assay.
For titration using plaque assays, virus preparations were serially diluted in complete DMEM and used to infect VeroE6 cells that were plated the day before in 24-well plates (200,000 cells/well). At 2 h postinfection, the inoculum was removed, and 1 ml of serum-free MEM containing 1.5% carboxymethyl cellulose was added. Cells were incubated at 37°C for 5 days. The cells were then fixed for 2 h with formaldehyde (5% final concentration). The cells were then extensively rinsed with water and stained with 1% crystal violet–10% ethanol for 15 to 30 min. After extensive washing with water, the plaques were counted, and the titers were calculated by taking into account the corresponding dilution factor.
Immunoblot analyses.
Samples were denatured in 2× protein sample buffer (200 mM Tris [pH 8.8], 5 mM EDTA, 0.1% bromophenol blue, 10% sucrose, 3% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol) and incubated for 5 min at 95°C. The proteins were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes by using a Mini-SDS-PAGE wet blotting apparatus (Bio-Rad, Munich, Germany). Membranes were blocked with 5% bovine serum albumin (BSA) in Tris-buffered saline–0.5% Tween 20 (TBST) and then incubated with primary antibodies overnight at 4°C. After three washes with TBST, the membranes were incubated with secondary horseradish peroxidase-conjugated antibodies, developed with the Western Lightning Plus-ECL reagent (Perkin-Elmer, Waltham, MA), and bands were imaged using an Intas ChemoCam Imager 3.2 (Intas, Göttingen, Germany). Signal intensities were quantified by using Intas LabImage 1D software.
RNA isolation and RT-qPCR analysis.
Total RNA was isolated from ∼106 Huh7 cells by using a NucleoSpin RNA II kit according to the instructions of the manufacturer (Macherey-Nagel, Düren, Germany). Purified RNA was eluted with 60 μl of double-distilled RNase-free water, quantified by spectrophotometry, and stored at −80°C until further use. All reverse transcription-quantitative PCRs (RT-qPCRs) were carried out with a two-step real-time RT-PCR kit (Applied Biosystems). Reverse transcription and real-time PCR were carried out in separate reactions. RNA was reverse transcribed by using Multiscribe reverse transcriptase (Applied Biosystems), as recommended by the manufacturer, and random primers. cDNA synthesis was carried out in a TGradient thermocycler (Biometra, Göttingen, Germany) using the following settings: 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min. cDNAs were either directly used for real-time PCR or stored at −80°C. The final volume of the real-time PCR mix was 15 μl and contained the following components: 7.5 μl of 2× green dye master mix (Qiagen, Hilden, Germany), 1.5 μl of primer mix (5 μM each), 3 μl of double-distilled water, and 3 μl of cDNA. Reactions were performed on an ABI Prism 7000 Sequence detection system using the following settings: 95°C for 10 min followed by 40 cycles at 95°C 30 s, 55°C for 60 s, and 72°C for 60 s. For each cell-specific primer set, reactions were carried out in triplicates. The following primers were used (MWG-Biotech, Martinsried, Germany): p62 (forward primer, 5′-AGGCGCACTACCGCGAT-3′; reverse primer, 5′-CGTCACTGGAAAAGGCAACC-3′), GAPDH (forward primer, 5′-GAAGGTGAAGGTCGGAGT-3′; reverse primer, 5′-GGGTCTCCTCTAACCTCTAGTCCT-3′), and DENV (forward primer, 5′-CAATATGCTGAAACGCGAGAGAA-3′; reverse primer, 5′-CCCCATCTATTCAGAATCCCTGC-3′). For all genes, the ΔΔCT method was used to calculate the relative expression levels.
Immunofluorescence and confocal microscopy.
Huh7 cells or Huh7 cells stably expressing mCherry-LC3 or GFP-LC3 were seeded into 24-well-plates containing glass coverslips. At 36 h postinfection, the cells were washed twice with phosphate-buffered saline (PBS), fixed with PBS–4% paraformaldehyde (PFA) for 15 min at room temperature and permeabilized with digitonin (50 μg/ml) or 0.2% Triton X-100 in PBS for 15 min at room temperature. The permeabilized cells were washed twice with PBS and blocked with PBS containing 5% BSA and 10% goat serum for 20 min at room temperature. Primary antibodies were added for 1 h at room temperature, and the cells were washed three times with PBS and incubated with a 1:1,000 dilution of secondary antibody in PBS containing 5% BSA for 1 h in the dark. The cells were washed once with PBS, incubated for 1 min with a 1:5,000-diluted solution of DAPI-PBS, and immediately washed three times for 10 min with PBS. Finally, the cells were mounted on glass slides with Slow-Fade Gold antifade reagent (Invitrogen/Molecular Probes, Darmstadt, Germany). The cells were imaged on a Nikon TE2000-E inverted confocal microscope equipped with an Ultraview ERS spinning disk system and a Plan-Apochromat VC 100× lens (NA 1.4). An EMCCD camera was used to sequentially record channels by defining emission discrimination in the following order: 647/700 nm, 568/610 nm, 488/510 nm, and 405/440 nm (emission/excitation). The acquired images were processed using the ImageJ and the Fiji software package (34).
Quantification of autolysosome formation.
Huh7 mCherry-LC3 cells were infected with the DENV NGC strain (multiplicity of infection [MOI] of 5) for 36 h, fixed in 4% PFA for 10 min, and immunostained against the lysosome marker Lamp2. Regions of interest were marked manually, and images were analyzed in an automated manner by using a Fiji script as follows. First, noise was suppressed by Gaussian filtering (sigma, 2 pixels), before the local background was estimated by applying morphological opening with a circular disc filter (radius, 8 pixels). Second, this background was subtracted, and the structures were provisionally identified in each channel using an automated threshold (the “triangle” method). Structures were retained if their areas exceeded 10 pixels. ImageJ's “Find Maxima” command was then applied to identify peaks in the “spots” channel surviving the previous thresholding steps, and the numbers and proportions of these peaks, overlapping with detected regions in the “other” channel, were counted for each cell. The data were extracted from n ≥ 70 cells for DENV, under uninfected or infected conditions.
ImageStream image-based, high-content flow-cytometry. (i) Quantification of autophagic flux.
For sample processing, 1.8 × 104 or 2 × 105 stable GFP-LC3-expressing Huh7 cells were seeded into each well of a six-well plate (BD Biosciences, Falcon) and infected with DENV or mock inoculated. Further incubation of cells with drugs, chemicals, or dyes was conducted as specified in Results. After the indicated treatments, the cells were treated with trypsin, fixed in 4% PFA for 20 min, resuspended in PBS (Sigma), and analyzed using an ImageStream X system (Amnis, Seattle, WA). For image analysis, IDEAS software (Amnis) was used. Briefly, the population of in-focus, single cells was selected for analysis. Segmentation masks of single cells and intracellular GFP-LC3 vesicles were generated. From these masks, the cellular area and intensity fractions for autophagy vesicles (AVs) and the number of AVs were calculated for each cell. Autophagic flux was determined based on the mean steady-state and BafA1-mediated cumulative population responses, as described in the legends to Fig. 1 and 2. The means for flux measurements are represented as bar graphs and bubble plots. For bubble plots, the circle size corresponds to mean steady-state AV content for each condition and time point. For experiments correlating AV content and NS3 expression, the cells were permeabilized with 0.3% Triton X-100 for 10 min, blocked with 3% BSA, and stained with anti-NS3 antibody overnight. NS3 immunofluorescence measurements were scaled between 0 and 1, based on the minimum and maximum fluorescence intensities, for each experiment.
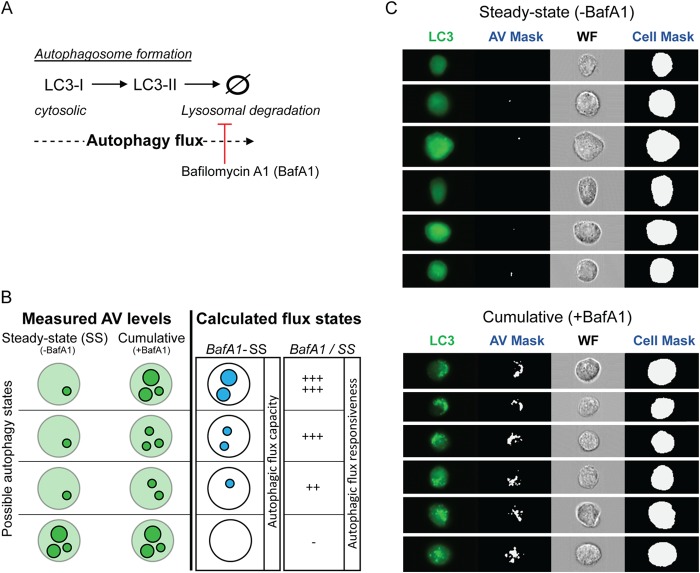
FIG 1.
Quantification of autophagic flux. (A) Schematic describing LC3 processing from cytosolic LC3-I to lipid-bound LC3-II, which imbeds on forming autophagosomes. LC3-II is degraded upon fusion with the lysosome. Formation and degradation of LC3-II-labeled autophagosomes constitutes autophagic flux. The lysosomal inhibitor bafilomycin A1 (BafA1) blocks degradation of autophagosomes by the lysosome. (B) Schematic illustrating used approaches to quantify autophagic flux in single cells. Different autophagy states of cells are based on the levels of autophagic vesicles (AVs). A steady-state level of AVs (SS) refers to the AV signal intensity in dimethyl sulfoxide (DMSO)-treated control cells, and BafA1 refers to the cumulative signal intensity of AVs due to blocked AV degradation. From steady-state and BafA1-treated conditions, we calculated the “autophagic flux capacity” by subtracting the steady-state AV levels from the BafA1-mediated cumulative AV levels (BafA1 − SS). “Autophagic flux responsiveness” refers to the ratio of AV signal intensity in BafA1-treated cells to AV signal intensity in untreated cells reflecting the steady-state (BafA1/SS). (C) Examples of image-based flow cytometry detection of GFP-LC3 expressing Huh7 cells, under full medium conditions, in the absence or presence of BafA1 (upper and lower panels, respectively). In addition, bright-field images of the steady-state and BafA1-mediated (i.e., cumulative) AV contents of single cells, collected at high resolution (40×), are shown. For each cell, masks for AVs and the cell were generated. Signal intensities of AVs and total cell GFP-LC3 fluorescence were used to calculate the fractional AV intensity (intensity AV/intensity cell). WF, wide field.
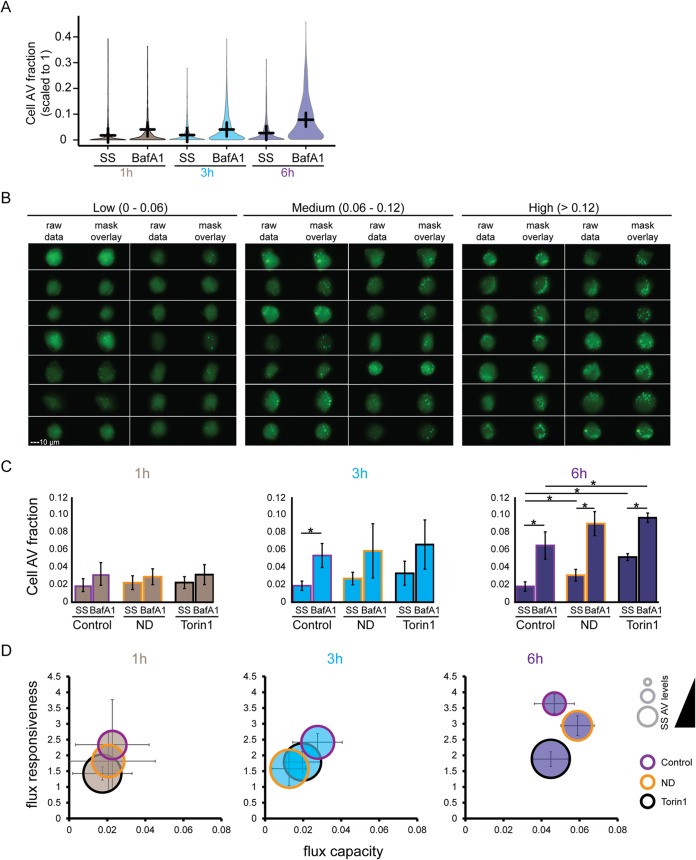
FIG 2.
ImageStream-based quantification of autophagic flux in Huh7 cells. (A) Time course cell population analysis of AVs in cells at steady-state (SS) and after BafA1 treatment. Huh7 GFP-LC3 cells were subjected to nutrient deprivation for 1, 3, and 6 h, in the absence (SS) or presence (BafA1) of 100 nM BafA1. Population distributions of cell AV fractions (see the calculation in Results) are shown. Lines indicate mean responses at indicated times. (B) Example heterogeneity of AV content in cells treated with BafA1 for 3 h in full medium. AV signal intensities were divided into three classes: low (0 to 0.06), medium (0.06 to 0.12), and high (>0.12) intensities. For each panel of a class, the fluorescence signal is shown on the left (raw data), and the AV mask overlays are shown on the right. (C) Bar graphs describing mean population responses of AV intensity fraction in the absence or presence of 100 nM BafA1 (SS or BafA1, respectively). Cells were additionally treated with Torin1 (2.5 μM) or cultured under nutrient deprivation (ND) or in full medium (Control). The data set represented here is an average of at least three independent experiments. The level of significance is denoted by an asterisk (*, P ≤ 0.05). (D) Significant block of autophagic flux in Huh7 GFP-LC3 cells after 6 h BafA1 treatment. Bubble plots represent calculated flux metrics as described in the legend to Fig. 1B. The sizes of the bubbles represent the steady-state AV levels. The data set represented here is an average of at least three independent experiments.
(ii) Quantification of endolysosomal activities.
DQ-BSA was used according to the manufacturer's protocol (Molecular Probes). Huh7 cells were treated with DQ-BSA (20 μg/ml) for 12 h at 37°C and then harvested for ImageStream analysis. The DQ-BSA signal was measured at a wavelength of 620 nm. Lysosomal (cathepsin B) activity was measured using Magic Red (MR; [Cresyl violet]MR-[RR]2; ImmunoChemistry Technologies, USA). Prior to harvesting, cells were treated with MR (20 μg/ml) during the last 2 h. Harvested cells were fixed in 4% PFA and subjected to ImageStream analysis. For image analysis, IDEAS software was used. Briefly, the population of in-focus single cells was selected for analysis. Segmentation masks of single cells and either DQ-BSA or MR-positive vesicles were generated. From these masks, the cellular area, vesicle intensity, and area fractions were calculated for each cell. Means for vesicle fraction intensities and vesicle sizes are represented as dot plots.
Statistical analyses.
Statistical analyses were performed as specified in the figure legends, and significance values were calculated by applying the two-tailed, unpaired Student t test.
RESULTS
Quantification of autophagic flux by image-based flow cytometry.
LC3 specifically associates with autophagosome membranes (35) and is the standard for monitoring autophagy through detection of its two processed forms: LC3-I, which is cytosolic, and LC3-II, which is bound to the autophagosomal membrane (Fig. 1A). To measure autophagic flux, steady-state LC3-labeled AV levels can be compared to AV levels accumulating upon inhibition of lysosomal activity with bafilomycin A1 (BafA1), termed cumulative AV levels (36, 37) (Fig. 1A and B). In the present study, we calculated from these two autophagy states two autophagic flux metrics. First, by subtracting steady-state AV levels from BafA1-induced AV levels (BafA1-treated − steady-state), we calculated an index of “autophagic flux capacity.” Second, from the ratio of cumulative to steady-state AVs (BafA1-treated/steady-state), we calculated the “autophagic flux responsiveness.” Thus, “autophagic flux capacity” reflects the amount of autophagosomes being formed and degraded in a given period of time, and “autophagic flux responsiveness” reports the fold change, which corresponds to sensitivity of a cell to autophagy induction for a given experimental condition.
Here, we sought to measure the autophagic flux in large populations of human hepatoma Huh7 cells stably expressing GFP-LC3, which we generated for this purpose, using the ImageStream system. This approach unites flow cytometry with high-resolution, fluorescence imaging (38), thus combining high sampling with spatial information obtained by high-resolution imaging, respectively. Therefore, the sensitivity and information content on autophagic flux are very much increased compared to standard flow cytometry (39–41). From each cell, an image was created, and segmentation masks for the cell and for AVs were generated and used to calculate the total cellular and vesicular GFP-LC3 fluorescence (see Fig. 1C for a representative set of images). The obtained values were then used to calculate the normalized fraction (scaled from 0 to 1) of GFP-LC3 localized within AVs for each cell within a population as follows: autophagy state of a single cell = cell AV fraction = AV fluorescence/total cell fluorescence.
Of note, LC3 is present on AVs of different maturation stages, and therefore GFP-LC3 punctae can represent pre-autophagosomal structures, autophagosomes, amphisomes or virus-induced autophagosome-like vesicles. All of these structures were considered in our autophagic flux measurements.
We first established the conditions for detecting autophagic flux in cell populations. Cells were subjected to nutrient deprivation (ND), in the absence or presence of BafA1 (100 nM), for 1, 3, and 6 h of treatment. As deduced from the cell AV fraction response, under steady-state condition (i.e., without BafA1 treatment), ND-mediated activation of autophagy resulted in small, nonsignificant changes for all time points (Fig. 2A). However, in BafA1-treated cells the AV levels increased with lengthened incubation period. Example images of cells kept in full medium and treated for 3 h with BafA1 revealed the accumulation of AVs falling into three phenotypic classes: low (0 to 0.06), medium (0.06 to 0.12), and high (>0.12) cell AV fractions (Fig. 2B), indicating the expected cell-to-cell heterogeneity due to differences in cell cycle or nutrient availability, for example.
To validate the approach further, we compared population means of basal flux under full medium conditions to the induction of autophagic flux by either nutrient deprivation or treatment with the specific mTOR inhibitor Torin1 (2.5 μM). The latter was used as positive control since it increases autophagosome production (42). These conditions, in the absence or presence of BafA1 (100 nM), were analyzed at 1, 3, and 6 h of treatment (Fig. 2C). At 1 and 3 h, BafA1 increased AV levels; however, only small differences were detected between basal (full medium) and activated (nutrient deprivation, Torin1) autophagy conditions. After 6 h of treatment, the AVs were significantly increased in cells treated with Torin1 or subjected to nutrient deprivation, relative to basal autophagy. Moreover, steady-state AV levels were increased by Torin1, relative to both full medium and nutrient deprivation conditions (Fig. 2C). By using calculated flux metrics to determine the time dependency of autophagic flux, we were able to distinguish population fold increases between all conditions and the population flux capacities between nutrient deprivation and Torin1 treatment (Fig. 2D). Thus, 6 h of cotreatment with BafA1 was required to significantly discriminate basal from activated autophagic flux in our cell system.
High-content imaging revealed that DENV blocked AV degradation and the induction of autophagosome formation.
We next infected Huh7 GFP-LC3 cells with DENV-2 (NGC strain) using an MOI of 5. Under these conditions, which we used for all subsequent experiments reported here, nearly 100% of cells were infected, as deduced from the detection of DENV nonstructural protein 3 (NS3) by immunofluorescence. Cells were infected for 30 h and then treated for 6 h with BafA1 (100 nM). Strikingly, relative to a mock-infected control, DENV-infected cells displayed increased levels of steady-state AVs (Fig. 3A and B). Consistent with these imaging results, Western blot analysis for LC3-I and -II levels in Huh7 cells revealed that after 36 h of infection, DENV increased the amounts of lipidated LC3 (LC3-II) under steady-state conditions (Fig. 3C).
FIG 3.
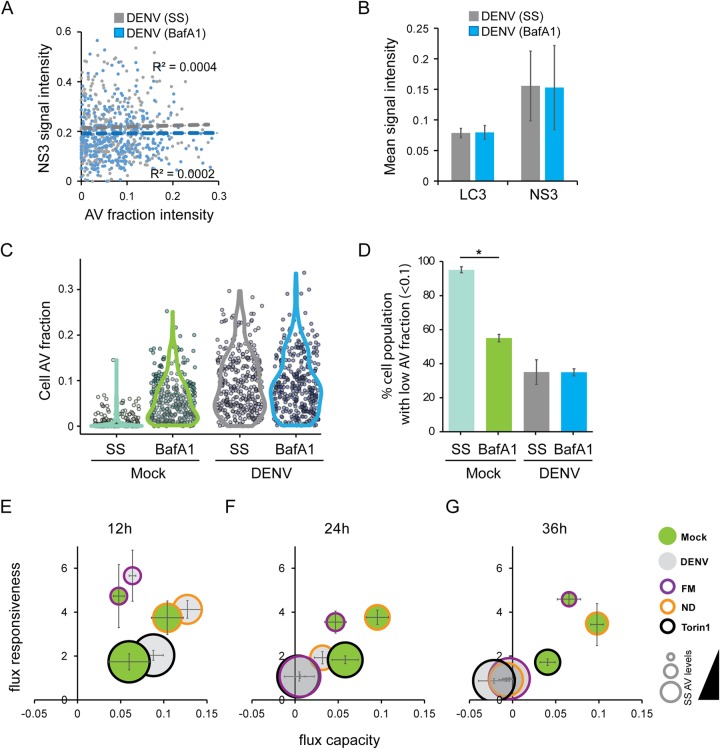
DENV impact on autophagic flux. (A) Examples of image-based flow cytometry detection of uninfected and DENV-infected Huh7 GFP-LC3 cells (upper and lower panels, respectively). Cells were mock infected or infected with the NGC isolate (MOI = 5). At 36 h postinfection, the cells were fixed and analyzed by ImageStream. For further details, see the legend to Fig. 1C. (B) Example of an immunofluorescence analysis of DENV-infected and uninfected Huh7/GFP-LC3 cells to validate infection rates. DENV NS3 was stained to visualize viral protein expression. (C) Measurement of endogenous LC3-II in untreated Huh7 GFP-LC3 cells (steady-state, SS) or BafA1-treated cells (BafA1). At 30 h after infection (NGC strain, MOI = 5), the cells were treated for 6 h with BafA1 (100 nM) or left untreated. The LC3-I and LC3-II levels were analyzed by Western blotting. In addition, LC3-II levels of three independent experiments were quantified by measuring the signal intensities with the INTAS software package and normalized to GAPDH. (D) Impact of BafA1 treatment on DENV replication kinetics. A subgenomic NGC replicon encoding a Renilla luciferase was electroporated into Huh7 cells. At 6 h before the indicated time points, the cells were treated with 100 nM BafA1 or a DMSO control. The cells were lysed, and the luciferase activity was measured. The data set represented here is an average of three independent experiments. (E to G) Bar graphs describing the mean population responses of the AV intensity fraction, the AV spot counts, and the AV area of Huh7 GFP-LC3 cells. (H) DENV blocks autophagic flux. Bubble plots represent calculated flux metrics as described in the legend to Fig. 1B. Bubble sizes represent steady-state (SS) AV levels. The data shown here represent the averages of three independent experiments.
To determine the impact of DENV on autophagic flux, we evaluated whether a block of autophagosomal degradation after 6 h BafA1 treatment would interfere with viral replication. We did not detect significant differences between control and BafA1-treated Huh7 cells that were electroporated with a subgenomic DENV NGC replicon encoding a firefly luciferase gene (Fig. 3D). Furthermore, in response to 6 h of BafA1 treatment, we observed a tendency for LC3-II to increase under full medium, Torin1, and nutrient depletion conditions, a finding consistent with a cumulative effect due to the BafA1-mediated inhibition of lysosomal degradation (Fig. 3C). In contrast, such accumulation was not seen in DENV-infected cells, arguing for a block of autophagic flux. For a more accurate quantification of this phenotype, we performed ImageStream analyses. We compared the signal intensities of AVs (i.e., the amount of GFP-LC3, which had accumulated with the vesicle), the number of AVs, and the AV areas (reflecting the sizes of vesicles). These parameters were compared between DENV-infected cells, cells with basal (control) autophagy, and cells in which autophagy was activated by either Torin1 treatment or nutrient deprivation. The mean population responses revealed that DENV infection resulted in increased levels of steady-state AVs, reaching levels that were equivalent to BafA1-mediated cumulative AV levels under conditions of activated autophagy (Fig. 3E to G). Based on the AV intensities, we calculated the autophagic flux, which clearly demonstrates that DENV induced a full block of the autophagic flux resulting in increased steady-state AVs (Fig. 3H).
We next examined the relationship between infection and the block to autophagy. For this purpose, we measured the viral infection state by immunodetection of DENV NS3 and correlated its content per cell with AV intensity levels by ImageStream analysis. No correlation was found under steady-state or BafA1 treatment conditions (Fig. 4A and B), suggesting that the block to autophagy is unrelated to the level of viral infection. Furthermore, we observed that in mock-inoculated cells, those with a low AV content massively responded to BafA1 treatment. In contrast, no population shift was detectable in DENV-infected cells (Fig. 4C). Even the low AV fraction (<0.1), which in mock-infected cells exhibits a high potential for further AV accumulation, did not respond to BafA1 treatment in DENV-infected cells, suggesting that AV content in DENV-infected cells is not maximal, thus implicating alterations to AV formation and/or degradation rates (Fig. 4D).
FIG 4.
Time-dependent influence of DENV infection on autophagic flux. (A) No correlation between NS3 and AV levels. Huh7 GFP-LC3 cells were infected with DENV (NGC isolate, MOI = 5) or mock inoculated. After 30 h, cells were either treated with DMSO control or 100 nM BafA1 for 6 h, followed by an immunostain against NS3 and ImageStream analysis. NS3 and normalized AV fraction intensities were plotted, and the intensities were tested for correlation. (B) Population mean AV fractions and NS3 signal intensities (scaled to 1) from three independent experiments, as described in panel A. (C) AV fraction shifts in cell populations. The experimental setup is as described for panel A. Population steady-state and BafA1-mediated cumulative AV fraction intensities at 36 h of mock or DENV infection are shown. Dots represent single cells, and the colored shape indicates the distribution of the population. (D) No shift of AV fraction in DENV-infected cells with high shift potential. The percentages of cells with a low AV fraction (<0.1) that respond to BafA1 treatment as described for panel C were determined. (E to G) Kinetics of autophagic flux during DENV infection. Time course population analysis of AV levels in mock-treated cells (steady-state, SS) and in cells treated with BafA1. Huh7 GFP-LC3 cells were infected with DENV NGC (MOI = 5) or mock treated for 12, 24, and 36 h. AT 6 h before harvesting, the cells were subjected to nutrient deprivation (ND), Torin1 (2.5 μM), or full medium (FM) conditions in the presence or absence of 100 nM BafA1. Bubble plots represent the calculated flux metrics, as described in the legend to Fig. 1B. The size of the bubble represents the steady-state AV levels (cells in the absence of BafA1). The data shown here represent the averages of three independent experiments.
Previously, the supporting role for autophagy in viral replication and lipid metabolism was detected during the first 24 h of infection (7). Therefore, we sought to determine the time point at which autophagy activity was disrupted by DENV. To do so, we measured basal (full medium) and activated (nutrient deprivation, Torin1) autophagic flux at 12, 24, and 36 h after infection. At 12 h of DENV infection, under all conditions, infected cells exhibited higher levels of autophagic flux compared to the corresponding mock-treated cells (Fig. 4E). In contrast, at 24 h postinfection, infected cells exhibited high steady-state AV levels under full medium and Torin1-treated conditions and fully inhibited autophagic flux. However, DENV-infected cells still exhibited autophagic flux in response to nutrient deprivation, albeit reduced compared to the control (Fig. 4F). At 36 h, DENV-infected cells exhibited high steady-state AV levels and a full block to autophagic flux under all conditions (Fig. 4G). These findings suggest a biphasic response of autophagy to DENV infection, where DENV infection activates (12 h) and then inhibits autophagy (at later time points). At 24 h postinfection, Torin1, which increases autophagosome production (42), had no impact on flux in DENV-infected cells. Basal autophagic flux was similarly blocked at this time point, whereas nutrient deprivation, which promotes lysosomal degradation of AVs, was still able to activate autophagic flux in DENV-infected cells. These findings suggest that DENV first blocks the formation of autophagosomes and then progressively reduces lysosomal AV degradation, which was complete by 36 h after infection.
Enhanced lysosomal activity in response to DENV infection.
In order to understand why AVs accumulate in DENV-infected cells, we next sought to investigate whether DENV infection affected lysosomal degradation capacity. Cathepsin B is a cysteine protease that belongs to a family of lysosomal proteases that are fully activated in the lysosomal low-pH environment (7). To measure protease activity, we utilized Magic Red (MR), a substrate that fluoresces upon cleavage by cathepsin B (Fig. 5A) (43). Cells were infected with DENV for 36 h and treated with MR for 2 h. Thereafter, ImageStream analysis was performed to calculate vesicle intensity, as well as vesicle area responses that were normalized to mock conditions. Although BafA1 treatment fully inhibited cathepsin activity, nutrient depletion resulted in a minor increase; in contrast, DENV increased cathepsin B activity >2-fold compared to uninfected cells (Fig. 5A). Thus, reduced autophagic flux by DENV infection did not correlate with a decreased lysosomal activity, arguing for an uncoupling between autophagosome and autolysosome formation.
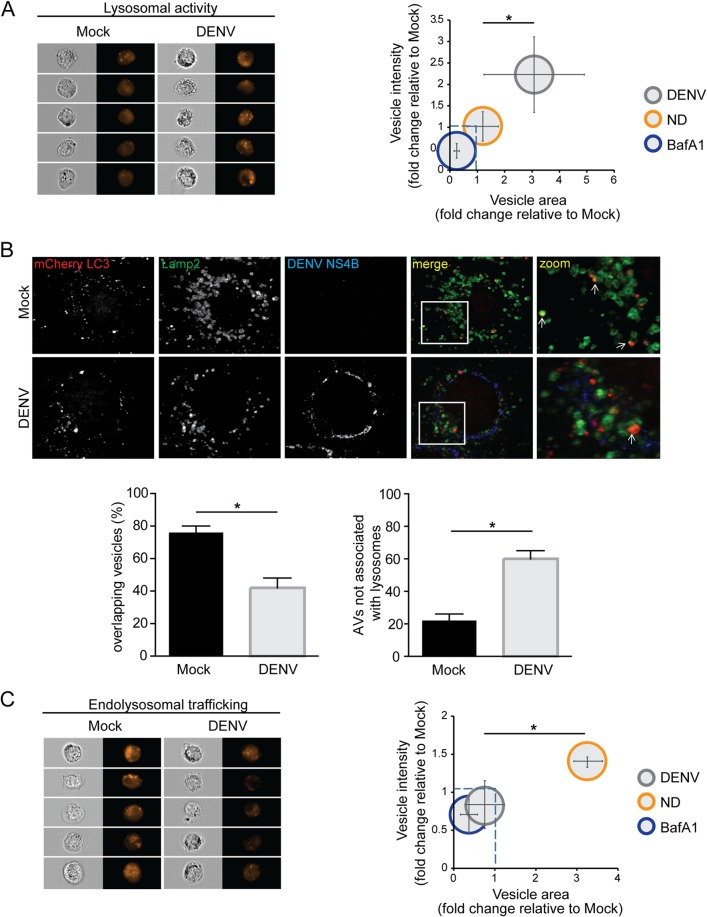
FIG 5.
Reduced autolysosome formation in DENV-infected cells. (A) Measurement of lysosomal activity in DENV-infected cells. Huh7 cells were mock infected or infected with DENV NGC (MOI = 5) for 36 h. In addition, mock-inoculated cells were subjected to nutrient deprivation, 100 nM BafA1, or full medium conditions for 6 h before harvesting. The cells were treated with Magic Red (20 μg/ml) for 2 h, fixed with 4% PFA, and further processed for flow cytometry-based image analysis. The data set represented here is an average of three independent experiments. The data were analyzed analogous to the description in the legend to Fig. 1B. (B) Colocalization of lysosomes with LC3-positive vesicles. Huh7 cells stably expressing the autophagic vesicle marker mCherry-LC3 were either mock treated or infected with DENV NGC (MOI = 5) for 36 h, respectively. The cells were fixed with 4% PFA and stained for lysosomes (Lamp2) and viral protein (NS4B). Samples were analyzed by confocal microscopy imaging. Images on the right represent magnifications of the regions highlighted in the corresponding merge images in the left. Note the colocalization of mCherry-LC3 punctae and lysosomes (marked with white arrow). Quantification of the total number of colocalizing spots in DENV-infected cells, as determined by signal overlap between mCherry-LC3 and the lysosomal marker Lamp2, was performed. For each condition, more than 50 cells were analyzed. The level of significance is measured by a Student t test and denoted as an asterisk (*, P ≤ 0.05). (C) Endolysosomal trafficking in DENV-infected cells. Huh7 cells were mock inoculated or infected with DENV NGC (MOI = 5) for 36 h. The cells were treated with DQ-BSA (20 μg/ml) for 12 h. Mock-inoculated cells were subjected to nutrient deprivation, 100 nM BafA1 treatment, or full medium conditions for 6 h before harvesting. The left panel shows examples of image-based flow cytometry detection of DQ-BSA; the right panel displays the relation of DQ-BSA vesicle areas and intensities. The data shown here represent the averages of three independent experiments.
DENV reduced endolysosomal trafficking and decreased autolysosome formation.
Based on these results, we explored the possibility that DENV alters the fusion of autophagosomes with lysosomes to form the degradative autolysosome. For optimum spatial resolution, required for colocalization analysis of autophagosomal and lysosomal markers, confocal microscopy was used. Huh7 cells stably expressing mCherry-LC3 were mock inoculated or infected with DENV for 36 h, and lysosomes were detected by using an antibody against the lysosomal marker Lamp2. Thus, in both uninfected and infected cells, autolysosomes were determined from the colocalization of autophagic vesicles (mCherry-LC3) with lysosomes (Lamp2). Analogous to earlier reports (7, 20), DENV NS4B did not colocalize with autophagosomal or lysosomal compartments (Fig. 5B). Quantitative image analysis revealed that DENV-infected cells exhibited a significant reduction in the fraction of LC3-positive vesicles overlapping with Lamp2-positive vesicles, compared to uninfected cells (Fig. 5B). Furthermore, in DENV-infected cells the number of autophagic vesicles not associated with lysosomes was increased, suggesting a block to fusion between autophagosomes and lysosomes (Fig. 5B).
We further investigated whether an altered potential of vesicle trafficking in DENV-infected cells could be the reason for the reduced autolysosome formation. Since both autophagy and endolysosomal pathways converge at lysosomes where intra- and extracellular material is degraded (44, 45), we determined the impact of DENV on endolysosomal trafficking. For this purpose, we used a BSA-derived fluorogenic substrate (DQ-BSA) (Fig. 5C) to measure endocytic trafficking of cargo to the lysosome (46). DQ-BSA is endocytosed and transported to the lysosome, where degradation decreases fluorophore self-quenching. The resulting increase of fluorescence is thus an indicator for endocytic flux and activity. Huh7 cells were left uninfected or were infected with DENV for 36 h and treated with DQ-BSA for 12 h prior to ImageStream analysis of vesicle intensity and area responses. Although nutrient deprivation increased the DQ-BSA response, DENV infection resulted in a decrease in DQ-BSA signal intensity compared to uninfected cells, suggesting that DENV infection strongly impairs endolysosomal trafficking to levels comparable to BafA1 inhibition of lysosomal activity (Fig. 5C), supporting the notion of a block to fusion between autophagosomes/endosomes and lysosomes (Fig. 5B and C).
DENV infection activates selective degradation of the p62 autophagy receptor.
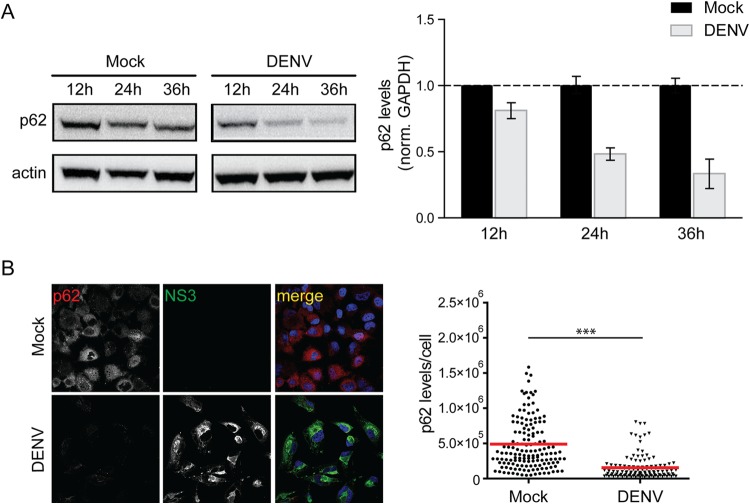
Given the global inhibition of autophagic flux, we sought to determine whether DENV might alter the abundance of the autophagy receptor p62, which is involved in autophagic vesicle maturation (47). Cells were infected with DENV and harvested at different time points after infection, and samples were analyzed by Western blotting (Fig. 6A) and immunofluorescence (Fig. 6B). Uninfected cells exhibited a moderate time-dependent decrease in p62 abundance (Fig. 6A), likely due to changes in cell confluence altering autophagy activity. In contrast, DENV infection resulted in a profound and time-dependent decrease in p62 protein levels. In comparison to uninfected cells, in DENV-infected cells p62 amounts were reduced by up to 65% at 36 h postinfection (Fig. 6A). Accordingly, similar results were obtained by quantitative image analysis 36 h after infection, revealing a 71% decrease of total p62 in DENV-infected cells (Fig. 6B).
FIG 6.
Impact of DENV infection on degradation of p62. (A) Time course analysis of p62 protein amounts. Huh7 cells were mock inoculated or infected with DENV NGC (MOI = 5) for given periods. Samples were harvested, and 40 μg of total cell protein was subjected to SDS-PAGE and Western blotting with p62- and β-actin (loading control)-specific antisera. p62- and β-actin-specific signals were quantified by using the ImageJ software package. The bar graphs on the right represent the p62 amounts relative to those detected in mock-inoculated cells at the indicated time points. (B) Immunodetection of p62 in DENV-infected cells. Huh7 cells were either mock treated or infected with DENV NGC (MOI = 5) for 36 h. The cells were fixed with 4% PFA and stained for the viral NS3 protein and p62. Samples were analyzed by confocal microscopy imaging. The scatter blot on the right shows the total p62-specific signal/cell. Signals were quantified by using the ImageJ software package. The level of significance is denoted by asterisks (***, P ≤ 0.001).
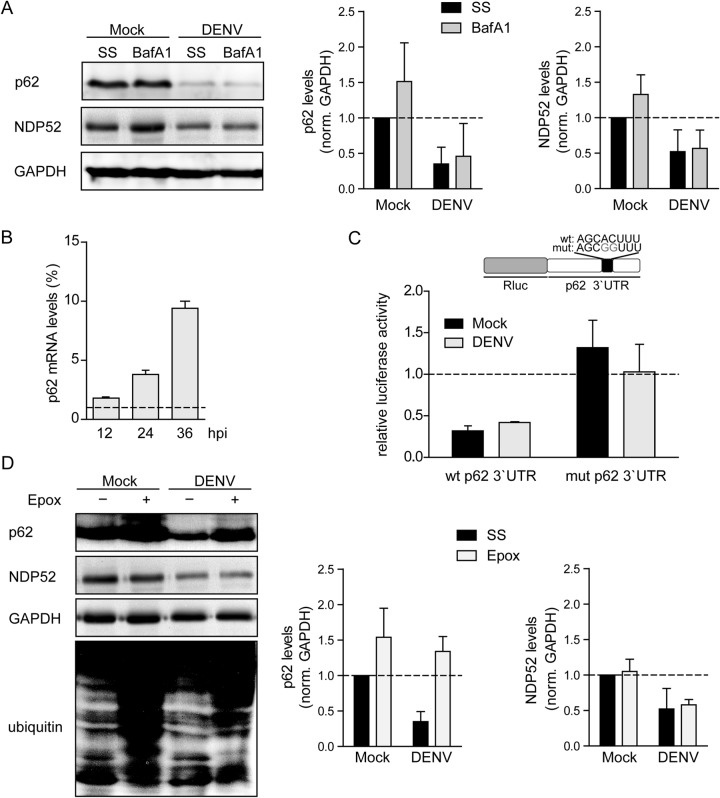
DENV-induced loss of p62 was surprising, since p62 is degraded by autophagy, which we determined to be progressively blocked upon DENV infection. We therefore compared the expression of p62 and another autophagic receptor, NDP52 (48), in mock- and DENV-infected cells, in the presence or absence of BafA1 treatment for 6 h. A 50% increase in p62 was detected in response to BafA1 treatment in uninfected cells compared to a negligible increase in DENV-infected cells, a finding that is consistent with a block of AV lysosomal targeting (Fig. 7A). NDP52 was also responsive to BafA1 in mock-treated cells, but the levels were unchanged in DENV-infected cells, further indicating a block to autophagy. Moreover, since BafA1 treatment did not rescue p62 expression in infected cells, DENV appears to modulate p62 protein amounts in a lysosome-independent manner.
FIG 7.
Enhanced proteasomal degradation of p62 in DENV-infected cells. (A) p62 is not degraded by the lysosome in DENV-infected cells. Huh7 cells were mock inoculated or infected with DENV NGC (MOI = 5). After 30 h, BafA1 (100 nM) was added for 6 h, or the cells were incubated in the absence of the drug (steady-state, SS). Samples were harvested, and 40 μg of total cell protein was subjected to SDS-PAGE and Western blotting with p62-, NDP52-, and GAPDH-specific antibodies. The bar graphs on the right display signal intensities that were quantified by using Intas quantification software to determine the levels of p62, NDP52, and GAPDH. Error bars represent the standard deviations (SD) of three independent experiments. (B) p62 RNA is upregulated in DENV-infected cells. Huh7 cells were infected with DENV NGC (MOI = 5) and harvested 12, 24, and 36 h after infection. Total RNA was purified and processed for RT-qPCR analysis to quantify mRNA amounts of GAPDH and p62. Values obtained with mock-inoculated cells were set to 1 and used for normalization. Error bars represent the SD of three experiments. (C) Translational repression of p62 is not enhanced in DENV-infected cells. A schematic of the luciferase reporter plasmids containing the full-length p62 3′UTR wild-type (wt) sequence or a mutated sequence containing two nucleotide substitutions in the seed sequence of the corresponding miRNA binding site (mut) is shown at the top. Each plasmid was cotransfected with a firefly luciferase control vector into Huh7 cells. After 24 h, the cells were mock inoculated or infected with DENV NGC (MOI = 5). After an additional 24 h, the cells were lysed, and the luciferase activity in cell lysates was quantified. The luciferase values were normalized to the luciferase 3′UTR (set to1) and to the firefly transfection control. Error bars represent the SD of three experiments. (D) p62 is degraded by the proteasome in DENV-infected cells. Huh7 cells were mock inoculated or infected with DENV NGC (MOI = 5). After 30 h, cells were treated for 6 h with epoxomycin (Epox; 100 nM) or mock treated (corresponding to steady-state, SS). Samples were harvested, and 40 μg of total cell protein was subjected to SDS-PAGE and Western blotting with p62-, NDP52-, GAPDH-, and ubiquitin-specific antibodies. Graphs on the right display the means signal intensities of three independent experiments, as quantified with the Intas software package, to determine the relative levels of p62, NDP52, and GAPDH.
We next determined whether reduced p62 amounts were due to altered mRNA levels. p62 mRNA was measured at 12, 24, and 36 h after DENV infection. Surprisingly, compared to the control, DENV induced p62 mRNA level up to 10-fold, indicating that mRNA abundance is not limiting for p62 protein expression (Fig. 7B). Since p62 expression is regulated by miRNAs encoded in the 17/20/93/106 cluster (49), we evaluated the possibility for suppression of p62 mRNA translation in DENV-infected cells. To this end, reporter constructs encoding the Renilla luciferase gene fused at the 3′ end to the full-length 3′ untranslated region (3′UTR) sequence of p62 of the wild-type sequence or containing mutations in the seed sequence binding site of the endogenous p62-targeting miRNA were generated (Fig. 7C). These constructs were transfected into Huh7 cells, together with a firefly luciferase control vector, in order to normalize the transfection efficiency. After 24 h of expression, the cells were infected with DENV, and both Renilla (Rluc) and firefly luciferase activities were determined 24 h later. In mock- and DENV-infected cells, the luciferase activity of the reporter containing the wild type, but not the mutant 3′UTR, was reduced compared to control Rluc mRNA, supporting a role for the miRNA 17/20/93/106 cluster in the basal regulation of p62 expression in Huh7 cells (Fig. 7C). However, only minor differences were observed between mock- and DENV-infected cells containing the wild-type p62 3′UTR reporter construct, suggesting that p62 mRNA translation is not altered in DENV-infected cells (Fig. 7C). These findings argue against a translational block to p62 by DENV infection.
As p62 can be degraded by the proteasome (50), we determined the impact of proteasomal degradation of p62 upon DENV infection. Huh7 cells were mock inoculated or infected with DENV and then, after 30 h, treated with the proteasome inhibitor epoxomycin (Epox) for 6 h, and the cell lysates were analyzed by Western blotting (Fig. 7D). In both mock- and DENV-infected cells, Epox increased the levels of ubiquitinylated proteins, demonstrating Epox-mediated inhibition of proteasome activity in both cell populations. Importantly, p62 levels were increased ∼50% in mock-inoculated cells and ∼300% in DENV-infected cells, arguing that proteasomal degradation might be the main mechanism of p62 depletion in DENV-infected cells. In contrast, the DENV-dependent decrease of NDP52 levels was insensitive to proteasomal, as well as lysosomal inhibition (Fig. 7D and A, respectively), arguing for a translational regulation in response to infection. In conclusion, these findings suggest that the loss of p62 in DENV-infected cells results from enhanced proteasomal degradation.
Novel role for p62 in the suppression of DENV replication.
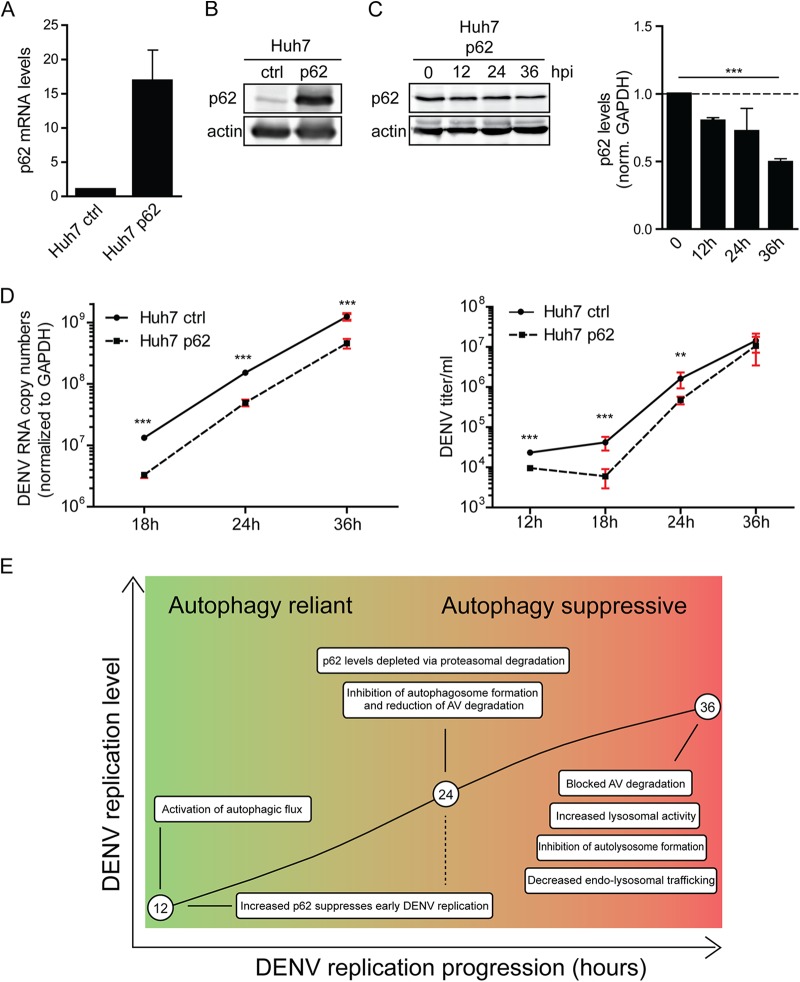
Since DENV induced the loss of p62, we reasoned that increased p62 abundance might interfere with viral replication. To test this hypothesis, we generated a Huh7 cell line stably overexpressing p62. Overexpression was confirmed at the level of p62 mRNA and protein (Fig. 8A and B). Importantly, while the levels of overexpressed p62 were also decreased over the course of DENV infection (Fig. 8C), the decline was attenuated compared to the one observed for endogenous p62, suggesting that, under these conditions, p62 levels exceed DENV-induced degradation capacity. We then determined the impact of increased p62 amounts on the DENV life cycle. Cells were infected with DENV (MOI = 0.25), and virus replication was determined by quantifying the amounts of intracellular viral RNA by RT-qPCR and infectious virus titers by plaque assay. DENV RNA was reduced 4-fold in response to p62 overexpression (Fig. 8D, left panel). Consistently, the titers of infectious DENV particles were significantly lower in response to p62 overexpression, although by 36 h after infection this phenotype leveled out (Fig. 8D, right panel). Overall, these findings show that high abundance of p62 inhibits DENV replication, arguing that virus-induced proteasomal degradation of this receptor and, more generally, autophagy inhibition are favorable to viral infection.
FIG 8.
p62 suppresses DENV replication. (A) Quantification of p62 mRNA overexpression. Huh7 cells stably overexpressing p62 or an empty vector control (ctrl) were generated by lentiviral transduction. p62 mRNA levels were quantified by RT-qPCR and normalized to GAPDH. (B) Quantification of p62 protein overexpression. A total of 40 μg of total cell protein was subjected to SDS-PAGE and Western blotting with p62- and β-actin-specific antibodies. (C) Degradation of overexpressed p62 in DENV-infected cells. Huh7 cells overexpressing p62 were infected with DENV NGC (MOI = 5) for the indicated periods, and lysates were analyzed by Western blotting for p62 and β-actin. The signal intensities were quantified by using Intas quantification software to determine the relative levels of p62 and β-actin. Error bars represent the SD of three independent experiments. (D) DENV RNA replication and virus production in p62-overexpressing cells. Huh7 cells overexpressing p62 or an empty vector control (ctrl) were infected with DENV NGC (MOI = 0.25) for the indicated periods. RNA was extracted and analyzed by RT-qPCR (left panel). Virus contained in cell culture supernatant was quantified by plaque assay on Vero cells (right panel). Error bars represent the SD of three independent experiments. The level of significance is denoted by asterisks (***, P ≤ 0.001). (E) Model for the differential impact of DENV on autophagic flux in the course of infection. For further details, see the text.
DISCUSSION
Autophagy activity can either promote or inhibit viral replication cycles (51–55). Here, we report findings that suggest a biphasic response of autophagy to DENV infection, where DENV infection initially activates and then later on inhibits autophagy. Although it has been reported by several groups that multiple components of the autophagy pathway are required for DENV replication (4, 6, 7, 27–29), we demonstrate here that DENV activates autophagy only during the early infection stage. Moreover, we established that, by 24 h after infection with the NGC strain, autophagosome formation is inhibited and lysosomal degradation of AVs is reduced. We determined that, by 36 h postinfection, the accumulation of AVs is the result of a block of lysosomal AV degradation. Surprisingly, we discovered that the autophagy receptor p62 was specifically depleted in DENV-infected cells via increased proteasomal degradation, and by overexpressing p62 we identified its role as a suppressor of DENV replication. Based on these systemic alterations of the autophagic flux, we propose a model how DENV manipulates autophagy during the course of infection (Fig. 8E): while autophagy activity is increased and supports DENV replication early during infection, at the later stage of infection specific autophagy suppression provides a viral replication advantage.
Fundamental to our work was the establishment of a quantitative approach to measure AVs under steady-state and BafA1 treatment conditions, from which we determined the impact of DENV infection on autophagic flux. This image-based flow cytometry analysis offers several advantages to conventional approaches. First, high sampling of cell populations permits accurate distribution measurements and high-confidence phenotype measurements. Second, the normalization of AV formation at the single-cell level permitted direct comparison among different conditions. Third, it is remarkable that under basal and activated conditions, steady-state AV levels were often comparable, and autophagic flux capacity in Huh7 cells was measurable only through a 6-h lysosomal inhibition by BafA1. These findings reinforce the concept of autophagy as a dynamic process and suggest that autophagy needs to be analyzed in a quantitative and time-resolved manner (7, 8, 17, 24). Our findings suggest that the accumulation of AVs was at least in part due to the block to lysosomal degradation and, beginning 24 h postinfection, DENV has already reduced autophagy induction capacity, at least in case of the NGC strain used here. This interpretation is consistent with a previous study that reported GFP-LC3 colocalization with LysoTracker Red (7), arguing that lysosomes were not sufficiently acidic to quench GFP fluorescence (30). Therefore, based on our findings, we suggest that AV accumulation is caused by a perturbation to AV turnover by DENV-2 infection. Although RNAi and Atg5 knockout studies clearly implicate autophagy as a DENV replication-promoting process, the results obtained using PI3K inhibitors are open to interpretation. 3MA has been used to show that autophagy induction is required for efficient virus production. However, 3MA suppresses autophagy induction by nutrient deprivation, whereas under typical growth conditions 3MA activates autophagy (56). Furthermore, inhibition of autophagy with the drug spautin-1 leads to increased production of noninfectious DENV particles (28), suggesting a role of autophagy during infectious particle production and/or release. The block of lysosomal targeting may be required by DENV for furin-mediated maturation of viral particles in a post-Golgi compartment. Alternatively, the excessive amount of DENV particles present in the cytoplasm may affect the general vesicular trafficking, hence disrupting AV lysosomal targeting. However, this remains controversial since in another study, 3MA and knockdown of Beclin1 expression had no impact on virus assembly while impairing RNA replication (7). In the present study, using the mTOR inhibitor Torin1, we demonstrate that autophagy induction is fully blocked in DENV-infected cells, suggesting that the virus renders the cell insensitive/uncoupled from PI3K. Since spautin-1 also promotes degradation of Vps34 (57), which is a regulator not only for autophagy but also for endocytic trafficking and cytokinesis (58), PI3K signaling may have nonautophagy roles in DENV replication.
Autolysosomes are reported as potential DENV replication sites, at least for DENV serotype 3 (6). Notably, that study revealed a similar increase in LC3-II levels upon DENV-3 infection. It would be interesting to determine similarities and differences between the DENV serotypes. Of note, it is quite possible that DENV induces autophagy at the early time points of infection to favor RNA replication but later in the replication cycle interferes with lysosomal targeting of AVs to favor the production of infectious viral particles. The latter strategy was recently reported for parainfluenza virus type 3 (52).
When considering possible reasons why DENV blocks autophagy, we note that overexpression of p62 suppressed viral replication and virus production arguing for an antiviral role of this autophagy receptor. We observed that p62 levels were steadily decreased in infected cells and speculate that this reduction might be required to overcome a p62-mediated restriction. Further work is required to determine the exact mechanism of p62 degradation (50, 59) and whether signaling pathways regulated by p62, including NF-κB signaling, the antioxidant response, and pathogen recognition (60), play a role in DENV restriction. In addition, since DENV has been shown to rely on a selective form of autophagy (i.e., lipophagy) for replication (7), it is possible that reduction of p62 abundance alters general autophagy (47) or lipophagy (7). It is also conceivable that DENV impairs late steps of autophagy to co-opt the host machinery responsible for AV biogenesis (i.e., ATG5, ATG12, and ATG16L1) and to manipulate ER-derived membranes eventually required for the formation of DENV vesicles packets and convoluted membranes (2).
In conclusion, we conducted the first quantitative and time-resolved analysis of autophagy activities in DENV-infected cells. Our results reveal that during infection DENV first activates and then inhibits both general and selective autophagy, blocking autophagosome formation and degradation by lysosomes and reducing the levels of the autophagy receptor p62. Our findings suggest that during the DENV replication process, autophagy shifts from a virus-supporting to a virus-suppressing process. The kinetic control of activation and inhibition of a fundamental cellular process described here illustrates the complex interaction between a virus and its host cell in order to achieve maximal virus replication.
ACKNOWLEDGMENTS
We are grateful to our colleagues in the Molecular Virology unit in Heidelberg for helpful discussions. We thank A. Davidson, University of Bristol, Bristol, United Kingdom, for providing the original DENV NGC cDNA clone and S. Erkeland, Erasmus MC Rotterdam, Rotterdam, Netherlands, for providing the p62-luciferase reporter constructs. We thank A. Ruggieri for generating Huh7 GFP-LC3B cells, W. Fischl for provision of Dengue virus constructs, U. Herian for excellent assistance with cell culture, and F. Huschmand for IT support. We are also very grateful to the Nikon Imaging Center (University of Heidelberg) for training and providing access to their equipment. We especially thank U. Engel at the Nikon Imaging Center for assistance with microscope handling.
N.R.B. was supported through the BMBF-funded project Immunoquant 0316170C and the SBCancer project within the Helmholtz Alliance on Systems Biology funded by the Initiative and Networking Fund of the Helmholtz Association. Work in the group of R.B. was supported by a grant from the BMBF project Immunoquant (0316170C).
REFERENCES
- 1.Ang F, Wong AP, Ng MM, Chu JJ. 2010. Small interference RNA profiling reveals the essential role of human membrane trafficking genes in mediating the infectious entry of dengue virus. Virol J 7:24. doi: 10.1186/1743-422X-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackenzie JM, Jones MK, Young PR. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 4.Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF, Lin YS, Yeh TM, Liu CC, Liu HS. 2008. Autophagic machinery activated by dengue virus enhances virus replication. Virology 374:240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. 2009. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J Gen Virol 90:448–456. doi: 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- 6.Khakpoor A, Panyasrivanit M, Wikan N, Smith DR. 2009. A role for autophagolysosomes in dengue virus 3 production in HepG2 cells. J Gen Virol 90:1093–1103. doi: 10.1099/vir.0.007914-0. [DOI] [PubMed] [Google Scholar]
- 7.Heaton NS, Randall G. 2010. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLean JE, Wudzinska A, Datan E, Quaglino D, Zakeri Z. 2011. Flavivirus NS4A-induced autophagy protects cells against death and enhances virus replication. J Biol Chem 286:22147–22159. doi: 10.1074/jbc.M110.192500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamacher-Brady A. 2012. Autophagy regulation and integration with cell signaling. Antioxid Redox Signal 17:756–765. doi: 10.1089/ars.2011.4410. [DOI] [PubMed] [Google Scholar]
- 10.Boyle KBand Randow F. 2013. The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol 16:339–348. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Johansen T, Lamark T. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Zhou L, Zhang Y, Leng Y, Pei XY, Lin H, Jones R, Orlowski RZ, Dai Y, Grant S. 2014. Targeting SQSTM1/p62 induces cargo loading failure and converts autophagy to apoptosis via NBK/Bik. Mol Cell Biol 34:3435–3449. doi: 10.1128/MCB.01383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. 2010. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 14.Delgado MAand Deretic V. 2009. Toll-like receptors in control of immunological autophagy. Cell Death Differ 16:976–983. doi: 10.1038/cdd.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gannage Mand Munz C. 2010. MHC presentation via autophagy and how viruses escape from it. Semin Immunopathol 32:373–381. doi: 10.1007/s00281-010-0227-7. [DOI] [PubMed] [Google Scholar]
- 16.Dreux M, Chisari FV. 2010. Viruses and the autophagy machinery. Cell Cycle 9:1295–1307. doi: 10.4161/cc.9.7.11109. [DOI] [PubMed] [Google Scholar]
- 17.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. 2008. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol 82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sir D, Liang C, Chen WL, Jung JU, Ou JH. 2008. Perturbation of autophagic pathway by hepatitis C virus. Autophagy 4:830–831. doi: 10.4161/auto.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. 2009. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5:937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- 21.Guevin C, Manna D, Belanger C, Konan KV, Mak P, Labonte P. 2010. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology 405:1–7. doi: 10.1016/j.virol.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrivastava S, Raychoudhuri A, Steele R, Ray R, Ray RB. 2011. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology 53:406–414. doi: 10.1002/hep.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards AL, Jackson WT. 2012. Intracellular vesicle acidification promotes maturation of infectious poliovirus particles. PLoS Pathog 8:e1003046. doi: 10.1371/journal.ppat.1003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dreux M, Gastaminza P, Wieland SF, Chisari FV. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A 106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke PYand Chen SS. 2011. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest 121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu LW, Huang YL, Lee JH, Huang LY, Chen WJ, Lin YH, Chen JY, Xiang R, Lee CH, Ping YH. 2014. Single-virus tracking approach to reveal the interaction of dengue virus with autophagy during the early stage of infection. J Biomed Opt 19:011018. doi: 10.1117/1.JBO.19.1.011018. [DOI] [PubMed] [Google Scholar]
- 28.Mateo R, Nagamine CM, Spagnolo J, Mendez E, Rahe M, Gale M Jr, Yuan J, Kirkegaard K. 2013. Inhibition of cellular autophagy deranges dengue virion maturation. J Virol 87:1312–1321. doi: 10.1128/JVI.02177-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YR, Hu HY, Kuo SH, Lei HY, Lin YS, Yeh TM, Liu CC, Liu HS. 2013. Dengue virus infection induces autophagy: an in vivo study. J Biomed Sci 20:65. doi: 10.1186/1423-0127-20-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. 2011. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol 9:38. doi: 10.1186/1741-7007-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller S, Sparacio S, Bartenschlager R. 2006. Subcellular localization and membrane topology of the dengue virus type 2 nonstructural protein 4B. J Biol Chem 281:8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- 32.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res 42:3858–3863. [PubMed] [Google Scholar]
- 33.Kovaleva V, Mora R, Park YJ, Plass C, Chiramel AI, Bartenschlager R, Dohner H, Stilgenbauer S, Pscherer A, Lichter P, Seiffert M. 2012. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res 72:1763–1772. doi: 10.1158/1538-7445.AM2012-1763. [DOI] [PubMed] [Google Scholar]
- 34.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamacher-Brady A, Brady NR, Gottlieb RA. 2006. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem 281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 37.Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. 2008. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 4:849–850. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 38.George TC, Basiji DA, Hall BE, Lynch DH, Ortyn WE, Perry DJ, Seo MJ, Zimmerman CA, Morrissey PJ. 2004. Distinguishing modes of cell death using the ImageStream multispectral imaging flow cytometer. Cytometry A 59:237–245. [DOI] [PubMed] [Google Scholar]
- 39.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phadwal K, Alegre-Abarrategui J, Watson AS, Pike L, Anbalagan S, Hammond EM, Wade-Martins R, McMichael A, Klenerman P, Simon AK. 2012. A novel method for autophagy detection in primary cells: impaired levels of macroautophagy in immunosenescent T cells. Autophagy 8:677–689. doi: 10.4161/auto.18935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demishtein A, Porat Z, Elazar Z, Shvets E. 2015. Applications of flow cytometry for measurement of autophagy. Methods 75:87–95. doi: 10.1016/j.ymeth.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 42.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. 2012. Cysteine cathepsins: from structure, function, and regulation to new frontiers. Biochim Biophys Acta 1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. 2012. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I. 2012. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol 32:1733–1744. doi: 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fader CM, Sanchez D, Furlan M, Colombo MI. 2008. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 47.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. 2009. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 49.Meenhuis A, van Veelen PA, de Looper H, van Boxtel N, van den Berge IJ, Sun SM, Taskesen E, Stern P, de Ru AH, van Adrichem AJ, Demmers J, Jongen-Lavrencic M, Lowenberg B, Touw IP, Sharp PA, Erkeland SJ. 2011. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood 118:916–925. doi: 10.1182/blood-2011-02-336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aichem A, Kalveram B, Spinnenhirn V, Kluge K, Catone N, Johansen T, Groettrup M. 2012. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J Cell Sci 125:4576–4585. doi: 10.1242/jcs.107789. [DOI] [PubMed] [Google Scholar]
- 51.Chiramel AI, Brady NR, Bartenschlager R. 2013. Divergent roles of autophagy in virus infection. Cells 2:83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding B, Zhang G, Yang X, Zhang S, Chen L, Yan Q, Xu M, Banerjee AK, Chen M. 2014. Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production. Cell Host Microbe 15:564–577. doi: 10.1016/j.chom.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 53.Sir D, Ann DK, Ou JH. 2010. Autophagy by hepatitis B virus and for hepatitis B virus. Autophagy 6:548–549. doi: 10.4161/auto.6.4.11669. [DOI] [PubMed] [Google Scholar]
- 54.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol 82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wileman T. 2007. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu Rev Microbiol 61:149–167. doi: 10.1146/annurev.micro.57.030502.090836. [DOI] [PubMed] [Google Scholar]
- 56.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM. 2010. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem 285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Hao Y, Choi A, Ke H, Ma D, Yuan J. 2011. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thoresen SB, Pedersen NM, Liestol K, Stenmark H. 2010. A phosphatidylinositol 3-kinase class III subcomplex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res 316:3368–3378. doi: 10.1016/j.yexcr.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 59.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. 2004. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol 24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deretic V. 2012. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol 24:21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]