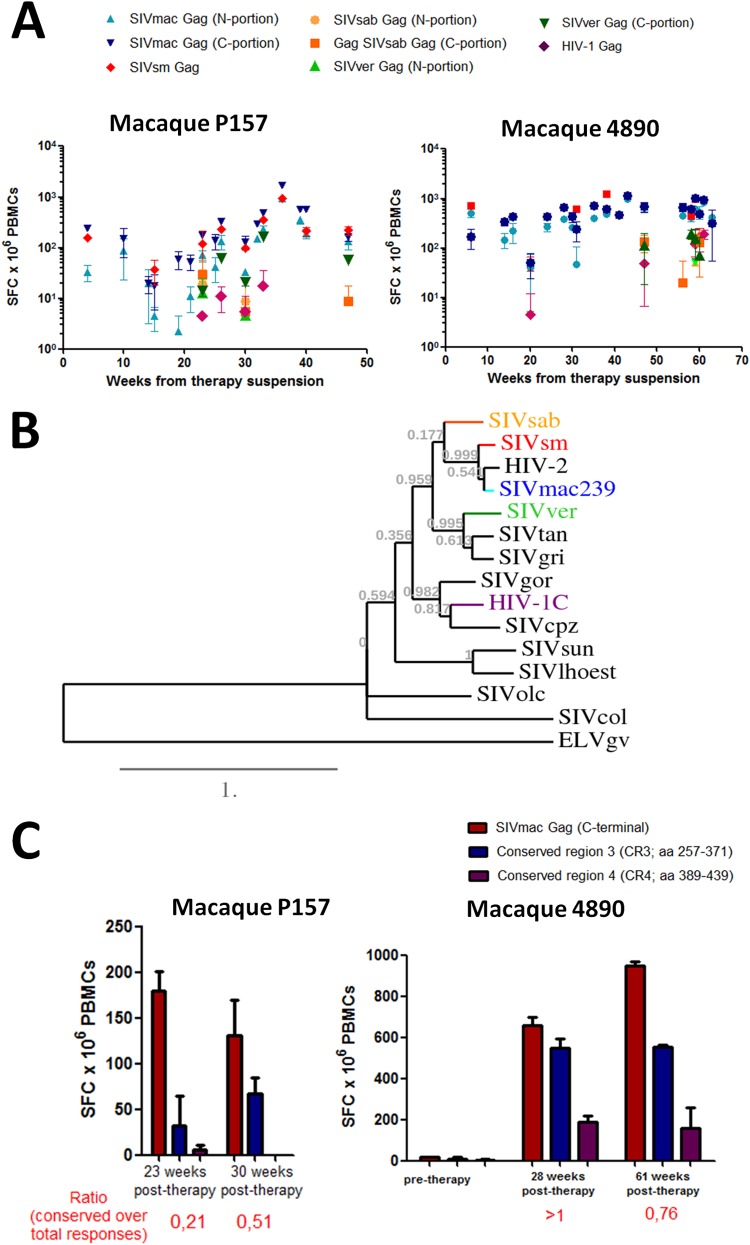
FIG 5.
Posttherapy development of broadly reactive anti-Gag immune responses in macaques treated with ART-auranofin-BSO. (A) Cell-mediated immune responses against the Gag antigens of various simian and human lentiviruses. The viruses employed are SIVmac239 (N portion and C portion, light and dark blue, respectively), SIVsm (red), SIVsab (N portion and C portion, ochre and orange, respectively), SIVver (N portion and C portion, light and dark green, respectively), and HIV-1 (purple). (B) Phylogenetic tree constructed on the basis of the Gag amino acid sequences of primate lentiviruses. Highlighted are the viruses used for the ELISpot analyses. Gag sequences from the following viruses were used to generate the tree: SIVsab, host Chlorocebus sabaeus (accession number U04005.1); SIVsm, host Cercocebus atys (accession number AF334679.1); HIV-2, host Homo sapiens (accession number A14062.1); SIVmac239, host Macaca mulatta (accession number AY611495.1); SIVver, host Chlorocebus aethiops (accession number M30931.1); SIVtan, host Chlorocebus tantalus (accession number U58991.1); SIVgri, host Chlorocebus pygerythrus (accession number M66437.1); SIVgor, host Gorilla gorilla (accession number FJ424871.1); HIV-1 (subtype C), host Homo sapiens (accession number AX457080.1); SIVcpz, host Pan troglodytes (accession number JN091691.1); SIVsun, host Cercopithecus solatus (accession number AF131870.1); SIVlhoest, host Cercopithecus lhoesti (accession number AF075269.1); SIVolc, host Procolobus verus (accession number AM713177.1); and SIVcol, host Colobus guereza (accession number AF301156.1). The recently sequenced Gag from ELVgv, an endogenous lentivirus from Galeopterus variegatus (order Dermoptera) (50) was used as an outgroup. The tree was constructed with phylogeny.fr (as described in reference 26) using a Clustal Ω alignment. (C) Cell-mediated immune responses against peptide pools encompassing conserved regions of the C-terminal half of the SIVmac239 Gag protein (i.e., CR3 and CR4; see Text S1 in the supplemental material). The sequences of the conserved regions employed for the analysis are shown in Text S1 in the supplemental material. Immune responses were measured by IFN-γ ELISpot. The ELISpot data are expressed as number of spot-forming cells (SFC) × 106 PBMCs. The ratio of conserved responses over the total is calculated, for each time point, as a sum of the mean number of SFC detected with CR3 and CR4 peptides divided by the mean number of SFC detected with peptides spanning the C-terminal half of the Gag.