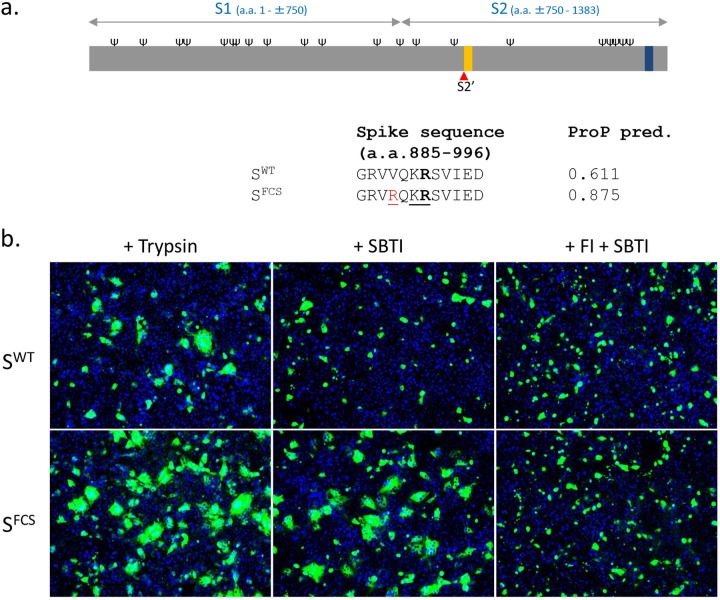
FIG 1.
The PEDV S protein with an artificial furin cleavage site at the S2′ position (SFCS) mediates trypsin-independent cell-cell fusion. (a) Schematic representation of the PEDV S protein (drawn to scale). Indicated are the positions of receptor binding domain S1 and membrane fusion domain S2, predicted N-glycosylation sites (Ψ; NetNGlyc server), the predicted S2′ cleavage site (red triangle), the fusion peptide (orange bar), and the transmembrane domain (black bar; TMHMM server). (Lower panel) Amino acid (a.a.) sequence at the putative S2′ cleavage site (R891, indicated in bold) for the wild-type CV777 S protein (SWT) and for a mutant S protein (SFCS) with a valine-to-arginine substitution at amino acid position 888 (colored in red), creating a furin cleavage site sequence (underlined) [R-X-(K/R)-R, where X is any amino acid]. Furin scores for the S2′ position predicted (pred.) by the ProP 1.0 server are indicated for both S variants. (b) Vero cells were transfected with expression plasmids encoding wild-type PEDV S (SWT) or SFCS and cultured in the absence or presence of a furin inhibitor (FI; 40 μM), a soybean trypsin inhibitor (SBTI; 40 μg/ml), or trypsin (15 μg/ml), as indicated. At 40 h posttransfection, cells were fixed and permeabilized. Nuclei (blue) were stained with DAPI (4′,6-diamidino-2-phenylindole), and PEDV S (green) expression was visualized by its C-terminally appended FLAG tag using anti-Flag monoclonal antibody (MAb) (Sigma). The transfection experiments were repeated two times, and representative images are shown.