ABSTRACT
There are 3 to 4 million new hepatitis C virus (HCV) infections annually around the world, but no vaccine is available. Robust T-cell mediated responses are necessary for effective clearance of the virus, and DNA vaccines result in a cell-mediated bias. Adjuvants are often required for effective vaccination, but during natural lytic viral infections damage-associated molecular patterns (DAMPs) are released, which act as natural adjuvants. Hence, a vaccine that induces cell necrosis and releases DAMPs will result in cell-mediated immunity (CMI), similar to that resulting from natural lytic viral infection. We have generated a DNA vaccine with the ability to elicit strong CMI against the HCV nonstructural (NS) proteins (3, 4A, 4B, and 5B) by encoding a cytolytic protein, perforin (PRF), and the antigens on a single plasmid. We examined the efficacy of the vaccines in C57BL/6 mice, as determined by gamma interferon enzyme-linked immunosorbent spot assay, cell proliferation studies, and intracellular cytokine production. Initially, we showed that encoding the NS4A protein in a vaccine which encoded only NS3 reduced the immunogenicity of NS3, whereas including PRF increased NS3 immunogenicity. In contrast, the inclusion of NS4A increased the immunogenicity of the NS3, NS4B, andNS5B proteins, when encoded in a DNA vaccine that also encoded PRF. Finally, vaccines that also encoded PRF elicited similar levels of CMI against each protein after vaccination with DNA encoding NS3, NS4A, NS4B, and NS5B compared to mice vaccinated with DNA encoding only NS3 or NS4B/5B. Thus, we have developed a promising “multiantigen” vaccine that elicits robust CMI.
IMPORTANCE Since their development, vaccines have reduced the global burden of disease. One strategy for vaccine development is to use commercially viable DNA technology, which has the potential to generate robust immune responses. Hepatitis C virus causes chronic liver infection and is a leading cause of liver cancer. To date, no vaccine is currently available, and treatment is costly and often results in side effects, limiting the number of patients who are treated. Despite recent advances in treatment, prevention remains the key to efficient control and elimination of this virus. Here, we describe a novel DNA vaccine against hepatitis C virus that is capable of inducing robust cell-mediated immune responses in mice and is a promising vaccine candidate for humans.
INTRODUCTION
Global efforts to generate an effective vaccine for hepatitis C virus (HCV) have been hampered, since correlates of sterilizing immunity have not been identified and traditional vaccine strategies have proved to be ineffective (1). Approximately 230 million individuals are infected, many of whom will develop serious liver disease, and since current treatment is costly and often results in serious side effects, this has limited the number of patients who are treated (2, 3). Much of the global effort to develop an effective HCV vaccine/treatment has been directed toward genotype 1 (gt1) (4, 5); however, gt3 is becoming more prominent (6) since it is detected more frequently in intravenous drug users (IDU) (7, 8) and is now the major genotype circulating in India and the United Kingdom (6, 9). Because there are genetic differences between gt1 and gt3 (10, 11) and assuming that initial HCV vaccines are likely to be genotype specific, greater focus on gt3 is warranted. Traditional vaccine methods that typically elicit neutralizing antibody response have been unable to generate protective immunity against HCV (12), resulting in a focus on strategies that generate cell-mediated immunity (CMI) (13–15). Since recovery from acute HCV infection and subsequent viral clearance require robust CMI that targets multiple HCV antigens (14, 16–20), this has resulted in a paradigm shift in vaccine design (21, 22). A successful vaccine that primarily induces CMI could function as a therapeutic vaccine by effective targeting of HCV-infected cells or as a prophylactic vaccine. Although HCV-specific CMI is unlikely to generate sterilizing immunity, an effective T-cell vaccine will prevent persistent infection and liver-associated disease, and since acute HCV infection is usually asymptomatic, this represents an acceptable, achievable objective. DNA vaccines generate CMI (23, 24), and several have been developed for use in animals (25–27). Furthermore, they are inexpensive, can be easily manufactured, and can be stored for long periods (28, 29), making them ideal for use in developing countries where the need for an effective HCV vaccine is greatest. Until now, strategies to enhance the immunogenicity of DNA vaccines have focused on encoding agonists of pathogen recognition receptors (PRRs), including Toll-like receptor (TLR) and NOD-like receptor agonists, or coinjection with TLR agonists such as poly(I·C) (30–33). These techniques target a specific PRR or a limited population of PRRs (32, 34). However, activation of a broader range of PRRs resulting from release of damage-associated molecular patterns (DAMPs) during natural lytic viral infection is able to produce a more robust immune response (31, 33, 35–37).
The rationale of the present study was to develop DNA vaccines that mimic the effect of live attenuated virus (LAV) vaccines which, although attenuated, are still lytic, resulting in activation of cell death pathways that are considered important to generate robust immunity (36, 37). Thus, the induction of necrosis in vaccine-targeted cells is expected to release the HCV immunogen and DAMPs, creating an adjuvant-rich milieu to enhance the HCV-specific immune responses after DNA immunization. Our results demonstrate that the induction of cell death in HCV antigen-positive cells, using a truncated, cytolytic form of the pore-forming protein, perforin (PRF) (38), significantly increased HCV-specific CD8+ T-cell responses relative to a canonical DNA vaccine. In addition, vaccination with DNA encoding a HCV polyprotein as an immunogen increased the breadth of the response to include all of the encoded antigens, without compromising the immunogenicity of the individual antigens.
MATERIALS AND METHODS
DNA vectors.
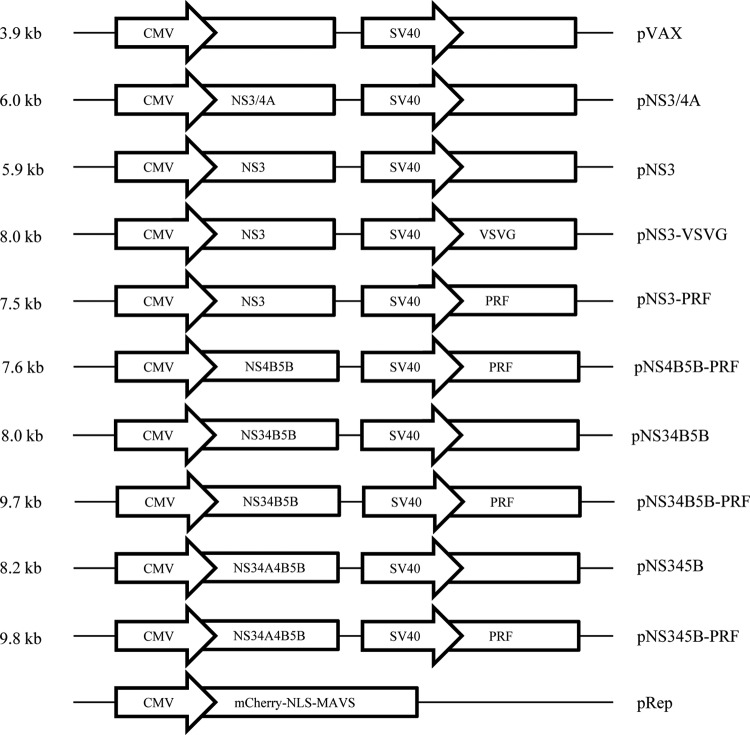
All constructs were created by standard methods, and a schematic diagram is shown in Fig. 1. The fluorescent reporter system (pRep), similar to that reported previously (39), kindly provided by Michael Beard, School of Molecular and Biomedical Science (Adelaide, South Australia, Australia), contained a fluorescent protein, mCherry, fused to a nuclear localization signal (NLS) and the C-terminal region of mitochondrial antiviral signaling (MAVS; amino acids [aa] 462 to 540) cloned into a pLenti backbone and purified using a PureLink HiPure plasmid midiprep kit (Life Technologies, Melbourne, Victoria, Australia). All DNA vaccine constructs were based on pVAX (Life Technologies). Codon-optimized genes (Gene Art, Germany) from HCV gt3a (GenBank accession number AF046866) were inserted downstream of the cytomegalovirus (CMV) promoter. An additional promoter, the simian virus 40 (SV40) promoter, and a poly(A)+ sequence were inserted into the pVAX backbone (40, 41), and genes for the cytolytic proteins, truncated perforin (PRF) lacking the final 12 residues of the C terminus (38), or vesicular stomatitis virus protein G (VSVG), were inserted downstream of this promoter. The VSVG clone was also kindly provided by Michael Beard. All DNA vaccine constructs were purified using a Qiagen (Doncaster, Victoria, Australia) endotoxin-free Mega kit.
FIG 1.
Schematic diagram of the bicistronic DNA vaccines used in the study. The DNA vaccine backbone, pVAX, was used to develop the following DNA vaccines: (i) HCV (poly)protein under the control of the CMV promoter and (ii) VSVG or PRF under the control of the SV40 promoter. An additional construct, which has a pLenti backbone, encodes mCherry, an NLS, and MAVS fused as a single protein under the control of the CMV promoter.
Cell culture.
HEK293T and Huh-7 cells were cultured in 24-well plates at 37°C in 5% CO2 in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and 1% penicillin-streptomycin and transfected with 800 ng of DNA using Lipofectamine LTX (Life Technologies) according to the manufacturer's protocol. Cell morphology posttransfection was assessed by light microscopy (using a Nikon Eclipse Ti microscope).
Western blot analysis.
Cells were harvested at 2 days posttransfection, lysates were prepared, and the concentrations of the proteins were determined using a Pierce BCA protein assay kit (Thermo Fisher Scientific, Inc., Scoresby, Victoria, Australia). The proteins were resolved by SDS–10% PAGE and transferred onto polyvinylidene difluoride membranes (Millipore, Kilsyth, Victoria, Australia). The membranes were blocked for 1 h in phosphate-buffered saline (PBS) containing 5% nonfat skim milk and 0.05% Tween 20 prior to incubation with primary antibodies overnight at 4°C. These primary antibodies included HCV gt3 patient pooled serum (kindly provided by Joseph Torresi, University of Melbourne, Melbourne, Victoria, Australia), rat anti-VSVG (Sigma-Aldrich, Castle Hill, New South Wales, Australia), rat anti-mouse PRF (Kamiya Biomedical Company, Seattle, WA), and mouse anti-β-actin (BD Biosciences [BD], North Ryde, New South Wales, Australia). After washing, the membranes were incubated for 1 to 2 h with horseradish peroxidase (HRP)-conjugated secondary antibodies (dilution 1:10,000) that included goat anti-human IgG-HRP, goat anti-mouse IgG-HRP, and goat ant-rat IgG-HRP (all Life Technologies). Finally, the proteins were visualized with a Western Lightning Ultra-ECL substrate chemiluminescence system (Perkin-Elmer, Melbourne, Victoria, Australia) according to the manufacturer's protocol using a Fujifilm LAS-4000 luminescent image analyzer.
Immunofluorescence.
At 2 days posttransfection with the DNA, HEK293T cells were fixed with 4% formalin (Sigma) for 20 min, permeabilized with methanol at −20°C, and then blocked in 2.5% bovine serum albumin (BSA) (Sigma) in PBS prior to overnight incubation with the primary antibodies at 4°C. The primary antibodies were HCV gt3 patient pooled serum and rat anti-PRF (Abcam, Melbourne, Victoria, Australia) were diluted in 1% BSA (Sigma) and 0.3% Triton X-100 (Sigma) in PBS. After a washing step, the cells were incubated for 1 h with fluorophore-conjugated secondary antibodies (1:300; goat anti-human-Alexa 555 or goat anti-rat-Cy5 [both from Life Technologies]) and then stained with DAPI (4′,6′-diamidino-2-phenylindole; Life Technologies). Cells were visualized by fluorescence microscopy (Zeiss LSM-700).
Animals and immunizations.
All experiments were approved by the University of Adelaide and the SA Pathology and Women's and Children's Health Network animal ethics committees. Six to eight-week-old C57BL/6 mice from the University of Adelaide Laboratory Animal Services were housed in HEPA-filtered individually vented cages. All interventions were performed under isoflurane or Domitor/Ketamine anesthesia (intraperitoneal [i.p.]) that was reversed by i.p. injection of Antisedan (Zoetis, West Ryde, New South Wales, Australia). Mice received various doses of endotoxin-free DNA in 50 μl injected into the dermal layer of the ear (intradermal injection) as described previously (40–43) and either two or three doses at 2-week intervals. At 2 weeks after the final vaccination, the mice were culled, and splenocytes were prepared as described previously (40, 41).
IFN-γ ELISPOT assay.
A gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISPOT) assay was performed on red blood cell-depleted splenocytes that were restimulated with 4-μg/ml portions of 15- to 19-mer, overlapping HCV gt3 peptides that were divided into three pools for NS3, two pools for NS4, and three pools for NS5B (BEI Resources, NIAID, Bethesda, MD), left unstimulated, or stimulated with phytohemagglutinin (PHA). Multiscreen-IP HTS plates (Millipore) were coated with anti-mouse IFN-γ (clone AN18; MabTech, Thomastown, Victoria, Australia), and secreted IFN-γ was detected with anti-mouse IFN-γ-biotin (clone R4-6A2; MabTech), streptavidin-AP, and SigmaFast BCIP/NBT (Sigma). Spots were counted automatically using an ELISPOT reader (AID Germany), and the number of spots in unstimulated splenocytes (∼0 to 50) subtracted from the number of spots in the peptide pool-stimulated splenocytes to generate the number of specific spot-forming units (SFU) per 106 cells was determined.
T-cell proliferation assay.
Proliferation of CD4+ and CD8+ T cells was assessed by flow cytometry after carboxyfluorescein diacetate succinimidyl ester (CFSE) staining. Briefly, splenocytes from vaccinated mice were labeled with 10 μM CFSE according to the CellTrace CFSE cell proliferation kit protocol (Life Technologies) and either stimulated in vitro with HCV peptides at 4 μg/ml representing the C-terminal region of NS3 (aa 422 to 631) or left unstimulated. After 4 days, CFSE-labeled splenocytes were stained with CD3-PerCP-Cy5.5, CD4-eFluor450, and CD8-APC-Cy7 (all from eBioscience, San Diego, CA), followed immediately by Hoechst staining prior to analysis by flow cytometry.
Flow cytometry.
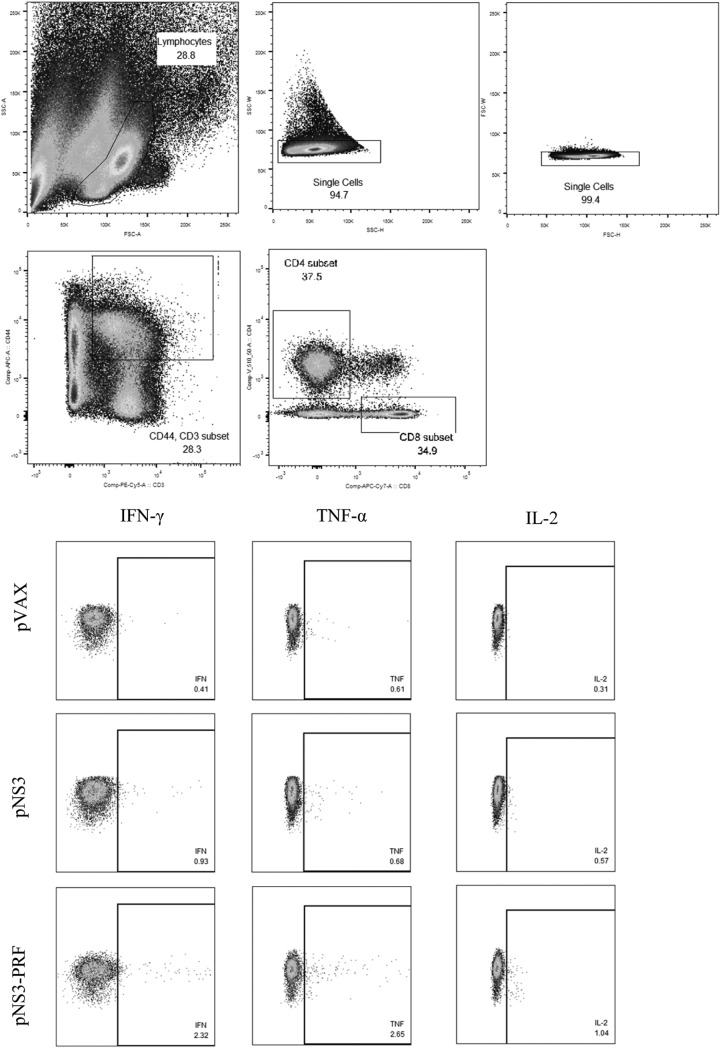
Multicolor intracellular cytokine staining (ICS) was performed on splenocytes stimulated in vitro with 4 μg/ml HCV peptides representing the C-terminal region of NS3 (aa 422 to 631) for 16 h or left unstimulated with the addition of a protein transport inhibitor (BD GolgiStop) for the final 12 h of stimulation. Staining was performed using BD fluorescence-activated cell sorting (FACS) Cytofix/Cytoperm and the following BD anti-mouse antibodies: CD3-PerCP-Cy5.5, CD8-APC-Cy7, CD44-APC, IL-2-FITC, IFN-γ-PE-Cy7, and TNF-α-PE. Cells were analyzed on a BD FACSCanto II flow cytometer using the gating strategy described in Fig. 2, and the results were analyzed by using FlowJo X.0.7 software (Ashland, OR).
FIG 2.
Gating strategy used to detect CD8 and CD4 T cells producing multiple cytokines in the splenocyte populations of vaccinated mice. Mice were vaccinated with three 10-μg doses or three 25-μg doses of DNA intradermally, and splenocytes were harvested on day 14 after the final vaccination for ICS. Cytokine profiles were determined by using flow cytometry. Splenocytes were gated on the lymphocyte population, followed by doublet discrimination, and then gated on CD3+ CD44+ cells and finally CD4+ or CD8+ cells to assess the frequency of IFN-γ, TNF-α, and IL-2. Representative blots for IFN-γ-, TNF-α-, and IL-2-positive cells are shown.
Statistical analysis.
Data are presented as means ± the standard errors of the mean (SEM). Statistical analysis was performed using unpaired Mann-Whitney tests, with P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***) considered significant. Analysis was performed using GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, CA).
RESULTS
Removal of NS4A increased NS3-specific immune responses.
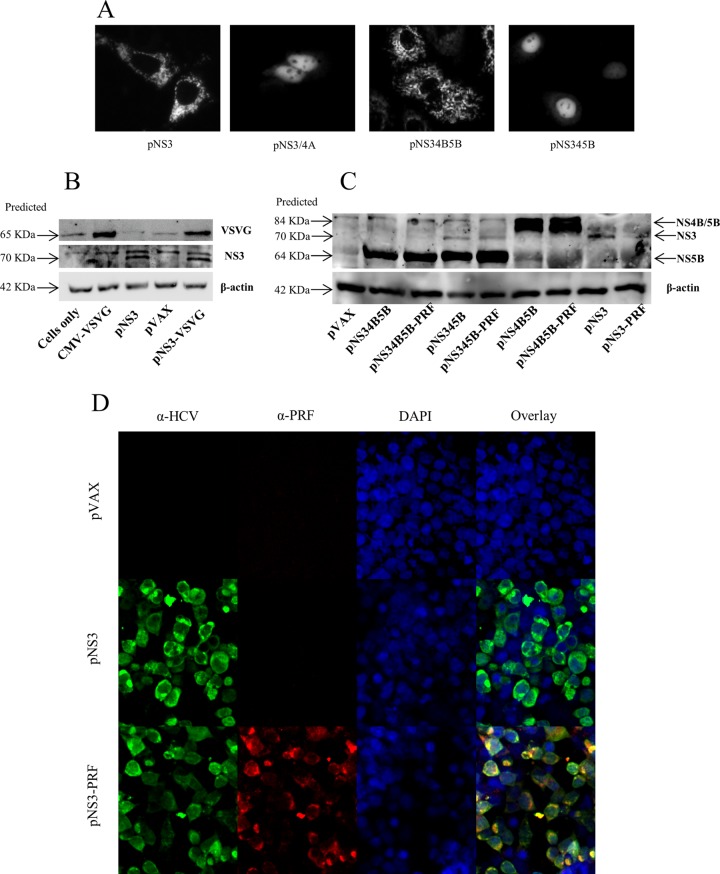
In addition to the cleavage of the HCV polyprotein, a functional NS3/4A protease also inhibits innate immune signaling via cleavage of MAVS and TIR-domain-containing adapter-inducing IFN-β (TRIF) (44–46). However, the HCV NS3 protease cofactor, NS4A, has also been reported to aid in protein stability (47). To determine whether NS3/4A expression would reduce the immunogenicity of NS3, plasmids encoding either only NS3 (pNS3) or both NS3 and 4A (pNS3/4A) (Fig. 1) were constructed. Protease activity associated with NS3 and NS3/4A was demonstrated using a previously described fluorescent reporter system (39, 48) (Fig. 1), in which a fluorescent protein, mCherry, is fused to a nuclear localization signal (NLS) and to amino acid residues 462 to 540 of MAVS that contain a cleavage site (C508) for the NS3/4A protease. Expression of mCherry-NLS-MAVS (pRep) is designed to result in punctate mitochondrial fluorescence, whereas cleavage at C508 will result in nuclear localization of mCherry-NLS. Cells cotransfected with pRep and pNS3 or pNS3/4A resulted in punctate cytoplasmic and nuclear expression of mCherry, respectively (Fig. 3A), confirming that NS3 had no protease activity in contrast to NS3/4A.
FIG 3.
HCV protein expression and protease activity. (A) To detect HCV protease cleavage of MAVS, Huh-7 cells were cotransfected with a plasmid (pRep) encoding a fluorescent reporter protein, i.e., mCherry, fused to an NLS and the C-terminal region of MAVS, as well as different constructs encoding various HCV proteins. At 24 h posttransfection, fluorescence was observed by fluorescence microscopy. The panels (left to right) show the intracellular localization of mCherry in cells cotransfected with pRep and either pNS3, pNS3/4A, pNS34B5B, and pNS345B, respectively. In general, cells which lack NS3/4A protease show punctate cytoplasmic fluorescence, whereas cells that express a functional NS3/4A protease show nuclear fluorescence. (B and C) Detection of HCV proteins in the lysates of transfected cells. HEK293T cells were transfected with DNA encoding NS3 with or without VSVG (B) and HCV proteins with or without PRF (C). At 48 h posttransfection, cell lysates were examined by Western blotting and probed with pooled HCV gt3 patient sera. Bicinchoninic acid and β-actin were used as loading controls. The predicted sizes of the HCV proteins are shown. Blots are representative of three independent experiments. (D) To detect perforin expression, HEK293T cells were transfected with plasmids in which perforin expression was controlled by the SV40 promoter. At 48 h posttransfection, the cells were fixed and permeabilized, and immunofluorescence was performed as described in Materials and Methods. The immunofluorescence images are representative of all perforin-expressing constructs.
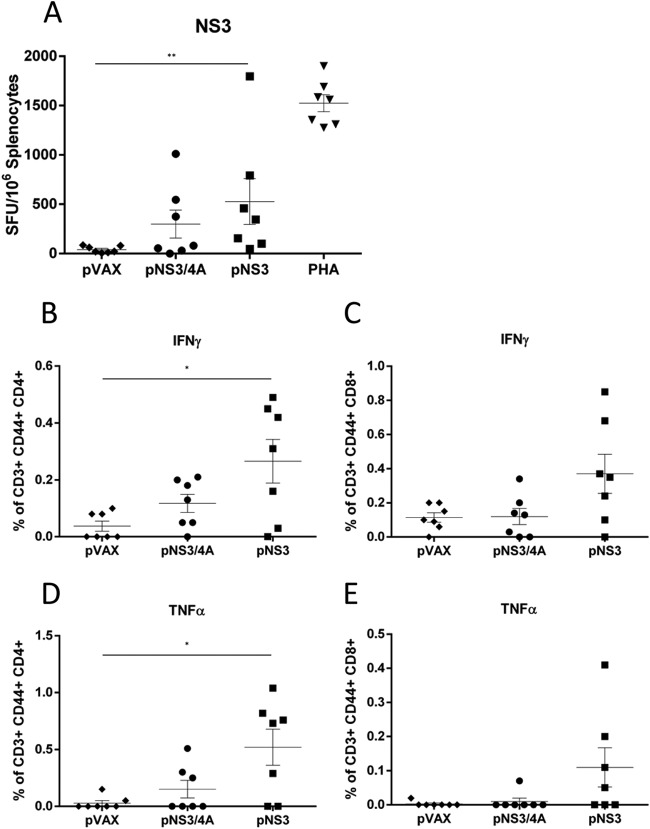
HCV protein expression was then confirmed by Western blotting (Fig. 3B and C). Since NS3/4A cleaves murine MAVS (49), we investigated the effect of NS4A expression in the context of NS3-specific immune responses in mice that were vaccinated with 3 doses of pNS3 or pNS3/4A. The CMI was assessed by IFN-γ ELISPOT assay to examine the breadth and magnitude of the responses, and ICS was performed to determine IFN-γ, tumor necrosis factor alpha (TNF-α), and interleukin-2 (IL-2) responses in HCV-specific CD4+ and CD8+ T-cell subsets. Mice vaccinated with pNS3 showed a significantly higher frequency (P = 0.0041) of SFU in NS3 peptide-stimulated splenocytes compared to splenocytes from mice vaccinated with the empty plasmid (pVAX) (528 v 42 SFU) (Fig. 4A). In comparison, mice vaccinated with pNS3/4A showed an intermediate response to NS3 that was not significantly different from that of mice vaccinated with pVAX (300 SFU versus 42 SFU) or mice vaccinated with pNS3 (300 SFU versus 528 SFU) (Fig. 4A). Since CD44+ is a marker of effector memory T (TEM) cells, which are important for recall responses (50), we examined the production of IL-2 from CD3+ CD44+ CD4+ TEM cells or from CD3+ CD44+ CD8+ TEM cells. There was no significant difference between the pVAX-, pNS3-, or pNS3/4A-vaccinated groups (data not shown), although the frequency of IFN-γ- and TNF-α-producing TEM cells was higher in pNS3-vaccinated mice than in pNS3/4A-vaccinated mice (Fig. 4B to E). Nevertheless, since NS3 appeared to be more immunogenic than NS3/4A, further experiments were performed using NS3 immunogen only.
FIG 4.
NS3 is more immunogenic than NS3/4A in C57BL/6 mice. Mice were vaccinated three times at 2-week intervals with the respective constructs, and splenocytes were harvested 14 days after the final vaccination. (A) Splenocytes were restimulated in duplicate, with overlapping peptides representing the complete HCV NS3 protein (gt3a), and IFN-γ secretion was measured by ELISPOT assay. The PHA control from the pVAX group is representative of all PHA-stimulated groups. The number of spots in unstimulated splenocytes was subtracted from the number in peptide-stimulated cells to generate the net number of NS3-specific SFU. The data are shown as the mean SFU/106 splenocytes ± the SEM. Cytokine profiles were determined using flow cytometry. Splenocytes were gated on CD3+, CD44+ cells and then CD4+ or CD8+ cells to assess the frequency of IFN-γ-producing CD4+ TEM cells (B), IFN-γ-producing CD8+ TEM cells (C), TNF-α-producing CD4+ TEM cells (D), and TNF-α-producing CD8+ TEM cells (E). Each symbol represents an individual mouse, and the data show the means (n = 7) ± the SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (Mann-Whitney test).
Coexpression of perforin enhances T-cell-mediated immune responses to NS3.
In order to mimic lytic virus cell death to increase antigen immunogenicity, the gene for a cytolytic protein was inserted into the constructs behind the weaker SV40 promoter. This combination provides the appropriate balance of immunogen expression and cell necrosis (41) and mimics the effect of LAV vaccines, which, although attenuated, are still lytic. Previous studies with a DNA vaccine have shown that intracellular expression of VSVG results in the development of syncytia, resulting in cell death (51, 52), and we have previously shown that the expression of PRF also results in cell death, although the mechanism was not defined (41, 43) but appeared to be nonapoptotic, i.e., necrotic (B. Grubor-Bauk et al., unpublished data). However, it has been reported that the truncated PRF variant fails to be exported from the endoplasmic reticulum and becomes highly toxic to the host cell (38). The expression of these proteins increased CMI to an immunogen that was expressed from a different plasmid in the case of VSVG (53, 54) or the same plasmid (PRF) (41, 43; Grubor-Bauk et al., unpublished). To determine whether expression of VSVG or PRF enhanced the immunogenicity of NS3, bicistronic vectors encoding both NS3 and either VSVG or PRF were constructed (Fig. 1), and protein expression was confirmed by Western blotting (Fig. 3B and C) or immunofluorescence (Fig. 3D). The latter experiment confirmed that NS3 and perforin were expressed in the same cells. Mice were vaccinated three times in a prime boost regimen, with an equimolar suboptimal dose of pVAX, pNS3, pNS3-VSVG, or pNS3-PRF (relative to 10 μg of the largest plasmid). Two weeks after the final boost, HCV-specific immune responses were measured by ELISPOT, ICS, and CFSE-based cell proliferation assays.
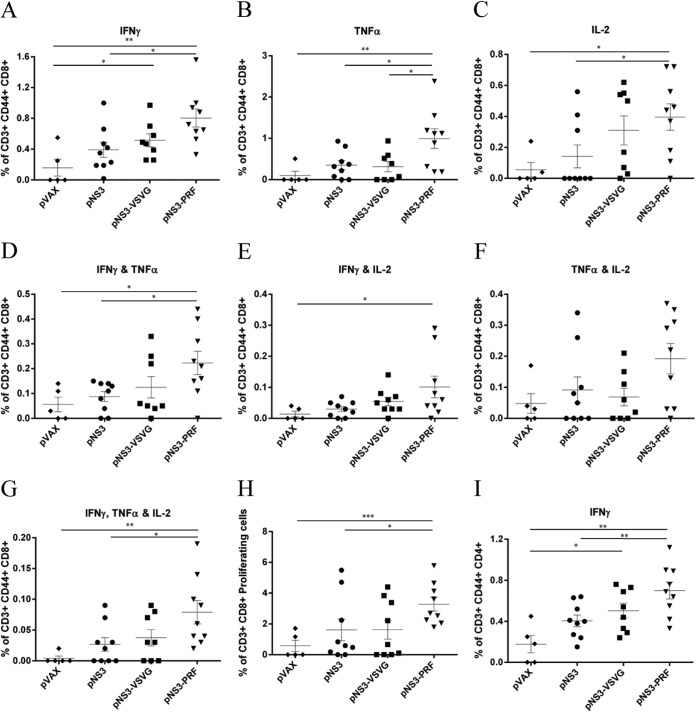
Mice vaccinated with pNS3-PRF showed significantly higher responses, as shown by an increased frequency of single cytokine producing (IFN-γ, TNF-α, or IL-2) CD3+ CD44+ CD8+ TEM cells compared to mice vaccinated with either pNS3 or pVAX (Fig. 5). However, pNS3-VSVG vaccinated mice only showed an increased frequency of IFN-γ-producing, but not TNF-α- or IL-2-producing, CD3+ CD44+ CD8+ TEM cells (P = 0.0241) compared to pVAX-vaccinated mice (Fig. 5A to C). Furthermore, vaccination with pNS3-PRF resulted in significantly higher IFN-γ and TNF-α double cytokine-producing CD3+ CD44+ CD8+ TEM cells compared to pNS3- or pVAX-vaccinated mice (P = 0.0129 and P = 0.0210, respectively) (Fig. 5D), whereas IFN-γ- and IL-2-producing CD3+ CD44+ CD8+ TEM cells from mice vaccinated with pNS3-PRF only were significantly increased compared to pVAX-vaccinated mice (P = 0.0310) (Fig. 5E). Although TNF-α- and IL-2-producing CD3+ CD44+ CD8+ TEM cells showed no significant differences between groups, pNS3-PRF-vaccinated mice showed the greatest response (Fig. 5F). In each of these analyses, mice vaccinated with pNS3-VSVG showed no significant increase in cytokine-producing cells relative to pVAX- or pNS3-vaccinated mice (Fig. 5D, E, and F). Finally, a significant increase in the frequency of multifunctional triple-cytokine-producing (IFN-γ, TNF-α, and IL-2) CD3+ CD44+ CD8+ TEM cells was noted in pNS3-PRF-vaccinated mice compared to pNS3- or pVAX-vaccinated mice (P = 0.0162 and P = 0.0015, respectively) (Fig. 5G), and although there was an increase in this cell population in the pNS3-PRF-vaccinated mice relative to the pNS3-VSVG-vaccinated mice, this was not significant.
FIG 5.
NS3 coexpressed with PRF is more immunogenic than NS3 alone. Splenocytes from vaccinated mice were stimulated with NS3 peptides, and cytokine production was analyzed using ICS and flow cytometry. Memory CD4+ and CD8+ T cells were gated to assess the frequency of IFN-γ-producing CD8+ TEM cells (A), TNF-α-producing CD8+ TEM cells (B), IL-2-producing CD8+ TEM cells (C), IFN-γ/TNF-α-double-producing CD8+ TEM cells (D), TNF-α/IL-2-double-producing CD8+ TEM cells (E), IFN-γ/IL-2-double-producing CD8+ TEM cells (F), IFN-γ/TNF-α/IL-2-triple-producing CD8+ TEM cells (G), and the proliferation of CFSE-labeled CD3+ CD8+ T cells (H). Splenocytes were labeled with CFSE and restimulated for 5 days with peptides representing the C-terminal third of NS3; the proliferation of CD3+ CD8+ T cells is shown as a percentage of total CD3+ CD8+ T cells. (I) Frequency of IFN-γ-producing CD4+ TEM cells after NS3 peptide stimulation. Each symbol represents an individual mouse, and the data show the means (n = 8 to 9) ± the SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (Mann-Whitney test).
Consistent with the data described above, cell proliferation studies, as a response to restimulation with NS3 peptides, showed increased proliferation of CD3+ CD8+ T cells from mice vaccinated with pNS3-PRF compared to CD3+ CD8+ T cells from pNS3- or pVAX-vaccinated mice (P = 0.0487 and P = 0.0010, respectively) but not pNS3-VSVG-vaccinated mice (Fig. 5H). pNS3-PRF-vaccinated mice also showed a significantly higher frequency of IFN-γ-secreting CD3+ CD44+ CD4+ TEM cells compared to mice vaccinated with pNS3 or pVAX (P = 0.0084 and P = 0.0040, respectively), while there was no difference in the frequency of CD3+ CD44+ CD4+ TEM cells secreting IL-2 or TNF-α (data not shown). Interestingly, pNS3-VSVG-vaccinated mice also showed a significant increase in the frequency of CD3+ CD44+ CD4+ TEM cells, but only relative to pVAX (P = 0.0287) (Fig. 5I). Thus, these data show that the coexpression of PRF but not VSVG resulted in enhanced immunogenicity of HCV NS3.
Efficient cleavage of HCV proteins improves immunogenicity when coexpressed with PRF.
Since a HCV vaccine should ideally generate responses against multiple antigens, we investigated whether multiple antigens could be used to increase the breadth of immunity against HCV without affecting the magnitude of the response against individual antigens. To address this point, we constructed several plasmids: pNS34B5B, pNS34B5B-PRF, pNS345B, and pNS345B-PRF (Fig. 1). These constructs encoded a truncated form of NS4B with an aa 1 to 84 deletion that has been reported to abrogate its immunosuppressive activity (55). The two polyproteins, NS34B5B and NS345B, contained the NS3/4A protease cleavage site between each individual protein, but the latter also contained NS4A to facilitate HCV polyprotein processing (Fig. 3C) (56, 57). However, since previous studies have shown that the NS polyprotein consensus cleavage sites other than NS4B/5A can be cleaved by NS3 alone (57, 58), it is highly likely that the consensus cleavage sites (D/E-X-X-X-X-C/T|S/A-X-X-X) in pNS34B5B, including the introduced 4B/5B site, was cleaved by NS3 alone. Thus, although the cleavage of NS3 was predicted from pNS34B5B, a mature NS3 protein was not detected (Fig. 3C, tracks 2 and 3), most likely because NS4A not only acts as a NS3 cofactor but also stabilizes NS3, since NS3 expressed in the absence of NS4A was reported to be promptly degraded (56). Our results are consistent with these early findings. Once protein expression (Fig. 3C) and MAVS cleavage were confirmed (Fig. 3A), the immunogenicity of these polyproteins was then examined to determine the effect of cleavage by NS3/4A.
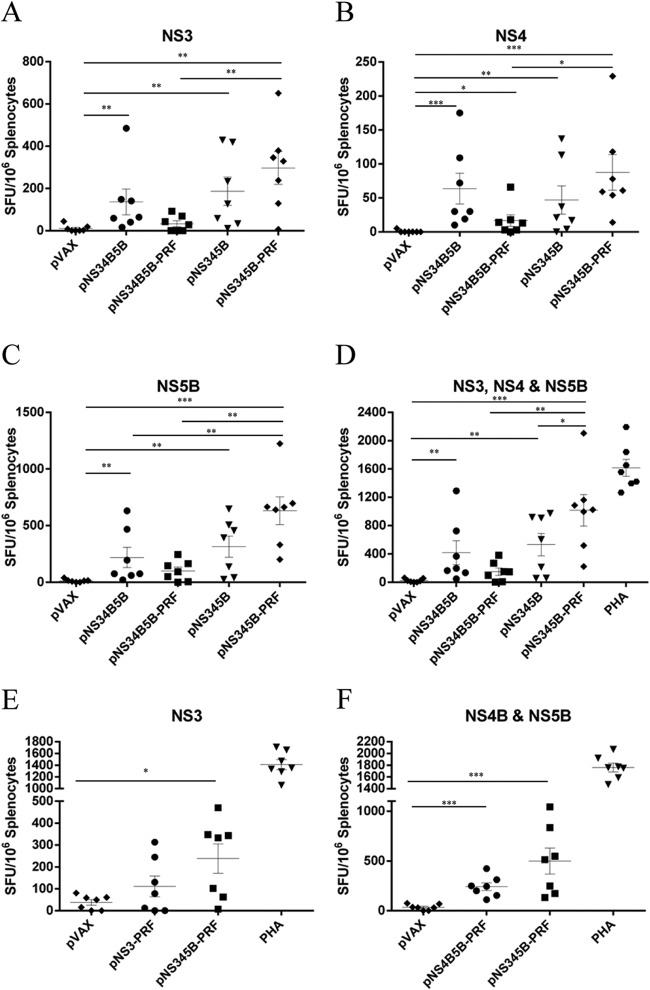
Mice were vaccinated twice with equimolar doses (50 μg for largest construct) of pVAX, pNS34B5B, pNS34B5B-PRF, pNS345B, or pNS345B-PRF (Fig. 1), and the immune responses were analyzed by IFN-γ ELISPOT assay. Mice vaccinated with either pNS34B5B or pNS345B showed similar levels of IFN-γ-producing cells when stimulated with NS3, NS4, or NS5B peptides (Fig. 6A to C). In contrast, the responses in mice vaccinated with pNS345B-PRF showed a significantly increased frequency of SFU (P = 0.0023) compared to pNS34B5B-PRF-vaccinated mice (1,016 SFU versus 151 SFU, respectively) (Fig. 6D), and mice vaccinated with pNS345B-PRF showed a significant increase (P = 0.0379) in SFU compared to pNS345B-vaccinated mice (1,016 SFU versus 532 SFU) (Fig. 6D). Somewhat surprisingly, pNS34B5B-PRF-vaccinated mice showed a reduced response compared to pNS34B5B-vaccinated mice; this represents the only occasion in recent years in which PRF has failed to induce increased immune responses (41, 43; Grubor-Bauk et al., unpublished). These data demonstrate that efficient processing of the polyprotein by the NS3/4A protease is required for optimal CMI when coexpressed with PRF.
FIG 6.
The adjuvant activity of PRF is retained in DNA vaccines encoding multiple HCV proteins. Mice were vaccinated twice with equimolar amounts of the plasmid DNA at 2-week intervals, and splenocytes from vaccinated animals were restimulated with overlapping peptides representing NS3 peptides (A), NS4 peptides (B), and NS5B peptides (C). (D) NS3, NS4, and NS5B combined analysis. (E and F) IFN-γ response in splenocytes from mice vaccinated with pNS345B-PRF or pNS3-PRF and restimulated with NS3 peptides (E) or with pNS345B-PRF or pNS4B5B and restimulated with NS4 and NS5B peptides (F). The PHA control from the pVAX group is representative of all PHA-stimulated groups. The number of spots in unstimulated splenocytes was subtracted from the number in peptide-stimulated cells to generate the net number of specific SFU. The data are shown as the mean SFU/106 splenocytes ± the SEM. Each symbol represents an individual mouse, and the data show the mean (n = 7) ± the SEM. *, P ≤ 0.05; ***, P ≤ 0.001 (Mann-Whitney test).
A vaccine encoding a polyprotein is as effective as the restricted antigen vaccine.
The above data indicated that pNS345B-PRF represented the optimum DNA vaccine candidate, since it elicited the strongest HCV-specific IFN-γ responses for all antigens encoded in the vaccine. To determine whether this multiantigen DNA vaccine elicited similar levels of HCV-specific CMI compared to vectors encoding single or limited HCV antigens, we vaccinated mice with pVAX, pNS3-PRF, pNS4B5B-PRF, or pNS345B-PRF. The mice received two equimolar doses (50 μg for the largest construct) 2 weeks apart, and the HCV-specific responses were then analyzed by an IFN-γ ELISPOT assay. Mice vaccinated with pNS345B-PRF showed higher responses against NS3 than mice vaccinated with pNS3-PRF (238 SFU versus 111 SFU) (Fig. 6E), and although this difference was not significant, there was a statistically significant difference in mice vaccinated with pNS345B-PRF compared to pVAX that was not apparent in the pNS3-PRF-versus-pVAX mice comparison. A higher response was also observed in pNS345B-PRF-vaccinated mice compared to pNS4B5B-PRF-vaccinated mice (499 SFU versus 242 SFU) (Fig. 6F), but again this was not significant.
Although the pNS345B-PRF vaccine appeared to be more effective than either pNS3-PRF or pNS4B5B, the increase in SFU was not significant. More importantly, this experiment proved that the multiantigen DNA vaccine elicits similar levels of cell-mediated immunity to the individual component antigens to that from DNA encoding a single or limited number of antigens.
DISCUSSION
The aim of this study was to increase HCV antigen immunogenicity by inducing death of vaccine-targeted cells, which is proposed to result in the release of antigens and DAMPs into the extracellular milieu. Released DAMPs will be sensed by immune cells, most importantly dendritic cells (DCs) (59), which will endocytose the HCV antigens that are then cross-presented via a nonclassical pathway (60).
DCs are essential for the generation of an adaptive immune response to an antigen (61, 62) and hence are critical targets for an effective vaccine (63); however, since these represent a rare population of cells, it has been challenging to target them directly with any vaccine. Our strategy offers a simple and yet elegant approach to indirectly target vaccine immunogens to DCs.
Previously, we noted that PRF was capable of increasing the immunogenicity of HIV-1 Gag and increased the frequency of CD11c+ CD8α+ DCs in the lymph nodes of vaccinated mice (41). It was also reported previously that VSVG induced cell death, increasing antigen immunogenicity (51, 52, 54), so we wanted to determine which of these proteins represented the more efficient adjuvant.
Before investigating the relative immunogenicity of HCV proteins coexpressed with PRF or VSVG, we sought to determine whether the NS3/4A protease activity affected the adaptive immune response. A previous study suggested that NS4A may have a beneficial effect on NS3 immunogenicity in mice (64). In contrast, we demonstrated that mice vaccinated with pNS3 elicited higher NS3-specific CMI responses than mice vaccinated with pNS3/4A (Fig. 4). This may have been due to NS3/4A cleavage of host proteins such as MAVS, which would reduce the overall immune response. Since pNS3-vaccinated mice elicited higher CMI responses than pNS3/4A-vaccinated mice, this construct was modified to also encode either VSVG or PRF to compare their adjuvant properties.
The cytolytic vaccine pNS3-VSVG showed modest enhancement of HCV-specific CMI in vaccinated mice compared to pNS3-vaccinated mice but was less effective than pNS3-PRF (Fig. 5). The pNS3-PRF vaccine induced significantly higher responses than did the canonical pNS3 vaccine, and an increased frequency was observed not only in single- and double-cytokine-expressing cells but also in multifunctional, triple-cytokine-producing CD8+ T cells that are highly desirable after vaccination and thought to be important for protective immunity (65) (Fig. 5). Indeed, correlates of protection have suggested that multifunctional CD8+ T cells are essential for effective clearance of HCV (17), as well as controlling other viral infections (66, 67). Furthermore, it has recently been demonstrated that a therapeutic human papillomavirus (HPV) DNA vaccine was able to clear the infection in seven of nine patients, with clearance correlating with polyfunctionality of CD8+ T cells (24). Thus, this provides a proof of concept that a therapeutic DNA vaccine which can elicit a robust polyfunctional CD8+ T-cell response could also result in elimination of persistent viral infection.
We then sought to demonstrate that the adjuvant effect of PRF not only increased CD8+ T-cell polyfunctionality but was equally effective in a multiple protein vaccine without compromising the immunogenicity of individual antigens. We compared the results of vaccination with pNS34B5B or pNS345B with or without PRF expression and noted that the inclusion of PRF in these vaccine constructs resulted in an increase in the immunogenicity of NS3, NS4, and NS5B after pNS345B-PRF vaccination, whereas their immunogenicity decreased after vaccination with pNS34B5B-PRF. Previous work with PRF has consistently increased the immunogenicity of single antigens (41–43; Grubor-Bauk et al., unpublished); hence, the reduced immunogenicity of NS34B5B in pNS34B5B-PRF vaccinated mice represents an unusual finding that is still unexplained. One possible explanation is that previous studies have shown that if the HCV polyprotein lacks NS4A, it has limited protease activity, thus resulting in reduced levels of individual proteins (56, 68). The addition of NS4A resulted in optimum cleavage of the polyprotein and hence an increase in the level of individual mature proteins (56, 68). It has also been demonstrated previously that long-lived, stable proteins are optimal for cross-priming in vivo compared to peptides or proteins that are readily degraded (69–72). Hence, the weakest response from pNS34B5B-PRF vaccination may have been due to HCV polyprotein instability.
Consequently, the DNA vaccine that encoded the NS345B polyprotein generated HCV-specific IFN-γ responses against the individual antigens that were higher than the responses elicited by constructs that encoded either NS3 or NS4B5B. Robust CMI responses against multiple HCV proteins, NS3 and NS5B in particular, are recognized as correlates of protection in patients who naturally clear HCV (14, 17, 18). Hence, a vaccine that is capable of inducing a robust CMI against NS3, NS4, and NS5B is pivotal for the development of protective immunity against HCV.
Our data demonstrated that PRF-induced death of HCV antigen-positive cells enhanced antigen immunogenicity and that multiple antigens, coexpressed with PRF in the same cell, benefited from this strategy. Overall, we have established a novel vaccine candidate that is capable of inducing a robust CMI response against a broad range of HCV proteins that warrants further development.
ACKNOWLEDGMENTS
We are grateful to Joseph Torresi for human anti-HCV serum and Michael Beard for the VSVG and pRep constructs. We also thank Stuart Howell for help with statistical analysis, the animal house staff, and past and present members of our laboratory for advice and technical support.
The following reagents were obtained through BEI Resources, NIAID, NIH: peptide array, hepatitis C virus, K3a/650, NS3 protein, NR-4066, NS4A protein, NR-4067, NS4B protein, NR-4068 and NS5B protein, NR-4070. This research was supported by grants APP1026293, 543139, and 543143 from the National Health and Medical Research Council (NHMRC) of Australia, grant BF040005 from the Australia-India Biotechnology Fund, and a grant from The Hospital Research Foundation (THRF). D.W. is supported by a fellowship from THRF.
REFERENCES
- 1.Swadling L, Klenerman P, Barnes E. 2013. Ever closer to a prophylactic vaccine for HCV. Expert Opin Biol Ther 13:1109–1124. doi: 10.1517/14712598.2013.791277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feeney ER, Chung RT. 2014. Antiviral treatment of hepatitis C. BMJ 348:g3308. doi: 10.1136/bmj.g3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shahid I, AlMalki WH, Hafeez MH, Hassan S. 2014. Hepatitis C virus infection treatment: an era of game changer direct acting antivirals and novel treatment strategies. Crit Rev Microbiol 6:1–13. doi: 10.3109/1040841X.2014.970123. [DOI] [PubMed] [Google Scholar]
- 4.Muir AJ. 2014. The rapid evolution of treatment strategies for hepatitis C. Am J Gastroenterol 109:628–636. doi: 10.1038/ajg.2014.66. [DOI] [PubMed] [Google Scholar]
- 5.Liang TJ. 2013. Current progress in development of hepatitis C virus vaccines. Nat Med 19:869–878. doi: 10.1038/nm.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. 2015. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thursz M, Fontanet A. 2014. HCV transmission in industrialized countries and resource-constrained areas. Nat Rev Gastroenterol Hepatol 11:28–35. doi: 10.1038/nrgastro.2013.179. [DOI] [PubMed] [Google Scholar]
- 8.Ampuero J, Romero-Gomez M, Reddy KR. 2014. Review article: HCV genotype 3: the new treatment challenge. Aliment Pharmacol Ther 39:686–698. doi: 10.1111/apt.12646. [DOI] [PubMed] [Google Scholar]
- 9.Sievert W, Altraif I, Razavi HA, Abdo A, Ahmed EA, Alomair A, Amarapurkar D, Chen CH, Dou X, El Khayat H, Elshazly M, Esmat G, Guan R, Han KH, Koike K, Largen A, McCaughan G, Mogawer S, Monis A, Nawaz A, Piratvisuth T, Sanai FM, Sharara AI, Sibbel S, Sood A, Suh DJ, Wallace C, Young K, Negro F. 2011. A systematic review of hepatitis C virus epidemiology in Asia, Australia, and Egypt. Liver Int 31(Suppl 2):S61–S80. doi: 10.1111/j.1478-3231.2011.02540.x. [DOI] [PubMed] [Google Scholar]
- 10.Rauch A, James I, Pfafferott K, Nolan D, Klenerman P, Cheng W, Mollison L, McCaughan G, Shackel N, Jeffrey GP, Baker R, Freitas E, Humphreys I, Furrer H, Gunthard HF, Hirschel B, Mallal S, John M, Lucas M, Barnes E, Gaudieri S. 2009. Divergent adaptation of hepatitis C virus genotypes 1 and 3 to human leukocyte antigen-restricted immune pressure. Hepatology 50:1017–1029. doi: 10.1002/hep.23101. [DOI] [PubMed] [Google Scholar]
- 11.Ripoli M, Pazienza V. 2011. Impact of HCV genetic differences on pathobiology of disease. Expert Rev Anti Infect Ther 9:747–759. doi: 10.1586/eri.11.94. [DOI] [PubMed] [Google Scholar]
- 12.Drummer HE. 2014. Challenges to the development of vaccines to hepatitis C virus that elicit neutralizing antibodies. Front Microbiol 5:329. doi: 10.3389/fmicb.2014.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roohvand F, Kossari N. 2012. Advances in hepatitis C virus vaccines, part 2: advances in hepatitis C virus vaccine formulations and modalities. Expert Opin Ther Pat 22:391–415. doi: 10.1517/13543776.2012.673589. [DOI] [PubMed] [Google Scholar]
- 14.Alvarez-Lajonchere L, Duenas-Carrera S. 2012. Complete definition of immunological correlates of protection and clearance of hepatitis C virus infection: a relevant pending task for vaccine development. Int Rev Immunol 31:223–242. doi: 10.3109/08830185.2012.680552. [DOI] [PubMed] [Google Scholar]
- 15.John M, Gaudieri S. 2014. Influence of HIV and HCV on T cell antigen presentation and challenges in the development of vaccines. Front Microbiol 5:514. doi: 10.3389/fmicb.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauer GM, Barnes E, Lucas M, Timm J, Ouchi K, Kim AY, Day CL, Robbins GK, Casson DR, Reiser M, Dusheiko G, Allen TM, Chung RT, Walker BD, Klenerman P. 2004. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology 127:924–936. doi: 10.1053/j.gastro.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Bharadwaj M, Thammanichanond D, Aitken CK, Moneer S, Drummer HE, Tracy S, Holdsworth R, Bowden S, Jackson D, Hellard M, Torresi J, McCluskey J. 2009. TCD8 response in diverse outcomes of recurrent exposure to hepatitis C virus. Immunol Cell Biol 87:464–472. doi: 10.1038/icb.2009.24. [DOI] [PubMed] [Google Scholar]
- 18.Urbani S, Amadei B, Fisicaro P, Tola D, Orlandini A, Sacchelli L, Mori C, Missale G, Ferrari C. 2006. Outcome of acute hepatitis C is related to virus-specific CD4 function and maturation of antiviral memory CD8 responses. Hepatology 44:126–139. doi: 10.1016/j.jhep.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Swadling L, Capone S, Antrobus RD, Brown A, Richardson R, Newell EW, Halliday J, Kelly C, Bowen D, Fergusson J, Kurioka A, Ammendola V, Del Sorbo M, Grazioli F, Esposito ML, Siani L, Traboni C, Hill A, Colloca S, Davis M, Nicosia A, Cortese R, Folgori A, Klenerman P, Barnes E. 2014. A human vaccine strategy based on chimpanzee adenoviral and MVA vectors that primes, boosts, and sustains functional HCV-specific T cell memory. Sci Transl Med 6:261ra153. doi: 10.1126/scitranslmed.3009185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes E, Folgori A, Capone S, Swadling L, Aston S, Kurioka A, Meyer J, Huddart R, Smith K, Townsend R, Brown A, Antrobus R, Ammendola V, Naddeo M, O'Hara G, Willberg C, Harrison A, Grazioli F, Esposito ML, Siani L, Traboni C, Oo Y, Adams D, Hill A, Colloca S, Nicosia A, Cortese R, Klenerman P. 2012. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci Transl Med 4:115ra111. doi: 10.1126/scitranslmed.3003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koup RA, Douek DC. 2011. Vaccine design for CD8 T lymphocyte responses. Cold Spring Harb Perspect Med 1:a007252. doi: 10.1101/cshperspect.a007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu MA. 2010. Immunologic basis of vaccine vectors. Immunity 33:504–515. doi: 10.1016/j.immuni.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Liu MA. 2011. DNA vaccines: an historical perspective and view to the future. Immunol Rev 239:62–84. doi: 10.1111/j.1600-065X.2010.00980.x. [DOI] [PubMed] [Google Scholar]
- 24.Kim TJ, Jin HT, Hur SY, Yang HG, Seo YB, Hong SR, Lee CW, Kim S, Woo JW, Park KS, Hwang YY, Park J, Lee IH, Lim KT, Lee KH, Jeong MS, Surh CD, Suh YS, Park JS, Sung YC. 2014. Clearance of persistent HPV infection and cervical lesion by therapeutic DNA vaccine in CIN3 patients. Nat Commun 5:5317. doi: 10.1038/ncomms6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao JC, Gregor P, Wolchok JD, Orlandi F, Craft D, Leung C, Houghton AN, Bergman PJ. 2006. Vaccination with human tyrosinase DNA induces antibody responses in dogs with advanced melanoma. Cancer Immun 6:8. [PMC free article] [PubMed] [Google Scholar]
- 26.Davis BS, Chang GJ, Cropp B, Roehrig JT, Martin DA, Mitchell CJ, Bowen R, Bunning ML. 2001. West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J Virol 75:4040–4047. doi: 10.1128/JVI.75.9.4040-4047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasuike M, Kondo H, Hirono I, Aoki T. 2007. Difference in Japanese flounder, Paralichthys olivaceus gene expression profile following hirame rhabdovirus (HIRRV) G and N protein DNA vaccination. Fish Shellfish Immunol 23:531–541. doi: 10.1016/j.fsi.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Saade F, Petrovsky N. 2012. Technologies for enhanced efficacy of DNA vaccines. Expert Rev Vaccines 11:189–209. doi: 10.1586/erv.11.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Saade F, Petrovsky N. 2012. The future of human DNA vaccines. J Biotechnol 162:171–182. doi: 10.1016/j.jbiotec.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwissa M, Nakaya HI, Oluoch H, Pulendran B. 2012. Distinct TLR adjuvants differentially stimulate systemic and local innate immune responses in nonhuman primates. Blood 119:2044–2055. doi: 10.1182/blood-2011-10-388579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timmermans K, Plantinga TS, Kox M, Vaneker M, Scheffer GJ, Adema GJ, Joosten LA, Netea MG. 2013. Blueprints of signaling interactions between pattern recognition receptors: implications for the design of vaccine adjuvants. Clin Vaccine Immunol 20:427–432. doi: 10.1128/CVI.00703-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgins SC, Mills KH. 2010. TLR, NLR agonists, and other immune modulators as infectious disease vaccine adjuvants. Curr Infect Dis Rep 12:4–12. doi: 10.1007/s11908-009-0080-9. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Q, Egelston C, Vivekanandhan A, Uematsu S, Akira S, Klinman DM, Belyakov IM, Berzofsky JA. 2008. Toll-like receptor ligands synergize through distinct dendritic cell pathways to induce T cell responses: implications for vaccines. Proc Natl Acad Sci U S A 105:16260–16265. doi: 10.1073/pnas.0805325105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Kang SM, Compans RW. 2009. Host responses from innate to adaptive immunity after vaccination: molecular and cellular events. Mol Cells 27:5–14. doi: 10.1007/s10059-009-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bridle BW, Clouthier D, Zhang L, Pol J, Chen L, Lichty BD, Bramson JL, Wan Y. 2013. Oncolytic vesicular stomatitis virus quantitatively and qualitatively improves primary CD8 T-cell responses to anticancer vaccines. Oncoimmunology 2:e26013. doi: 10.4161/onci.26013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks CL, Picker LJ, King CR. 2013. Development of replication-competent viral vectors for HIV vaccine delivery. Curr Opin HIV AIDS 8:402–411. doi: 10.1097/COH.0b013e328363d389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennan AJ, Chia J, Browne KA, Ciccone A, Ellis S, Lopez JA, Susanto O, Verschoor S, Yagita H, Whisstock JC, Trapani JA, Voskoboinik I. 2011. Protection from endogenous perforin: glycans and the C terminus regulate exocytic trafficking in cytotoxic lymphocytes. Immunity 34:879–892. doi: 10.1016/j.immuni.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 39.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol 28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrod TJ, Grubor-Bauk B, Gargett T, Li Y, Miller DS, Yu W, Major L, Burrell CJ, Wesselingh S, Suhrbier A, Gowans EJ. 2014. DNA vaccines encoding membrane-bound or secreted forms of heat shock protein 70 exhibit improved potency. Eur J Immunol 44:1992–2002. doi: 10.1002/eji.201343983. [DOI] [PubMed] [Google Scholar]
- 41.Gargett T, Grubor-Bauk B, Garrod TJ, Yu W, Miller D, Major L, Wesselingh S, Suhrbier A, Gowans EJ. 2014. Induction of antigen-positive cell death by the expression of perforin, but not DTa, from a DNA vaccine enhances the immune response. Immunol Cell Biol 92:359–367. doi: 10.1038/icb.2013.93. [DOI] [PubMed] [Google Scholar]
- 42.Garrod T, Grubor-Bauk B, Yu S, Gargett T, Gowans EJ. 2014. Encoded novel forms of HSP70 or a cytolytic protein increase DNA vaccine potency. Hum Vaccin Immunother 10:2679–2683. doi: 10.4161/hv.29527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gargett T, Grubor-Bauk B, Miller D, Garrod T, Yu S, Wesselingh S, Suhrbier A, Gowans EJ. 2014. Increase in DNA vaccine efficacy by virosome delivery and coexpression of a cytolytic protein. Clin Transl Immunol 3:e18. doi: 10.1038/cti.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, Wilkinson P, Julkunen I, Vitour D, Meurs E, Hiscott J. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol 80:6072–6083. doi: 10.1128/JVI.02495-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M Jr, Lemon SM. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A 102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morikawa K, Lange CM, Gouttenoire J, Meylan E, Brass V, Penin F, Moradpour D. 2011. Nonstructural protein 3-4A: the Swiss army knife of hepatitis C virus. J Viral Hepat 18:305–315. doi: 10.1111/j.1365-2893.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- 47.Wolk B, Sansonno D, Krausslich HG, Dammacco F, Rice CM, Blum HE, Moradpour D. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J Virol 74:2293–2304. doi: 10.1128/JVI.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eyre NS, Beard MR. 2010. Hepatitis C virus infection in living color. Hepatology 51:1852–1855. doi: 10.1002/hep.23695. [DOI] [PubMed] [Google Scholar]
- 49.Ahlen G, Derk E, Weiland M, Jiao J, Rahbin N, Aleman S, Peterson DL, Pokrovskaja K, Grander D, Frelin L, Sallberg M. 2009. Cleavage of the IPS-1/Cardif/MAVS/VISA does not inhibit T cell-mediated elimination of hepatitis C virus nonstructural 3/4A-expressing hepatocytes. Gut 58:560–569. doi: 10.1136/gut.2007.147264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oehen S, Brduscha-Riem K. 1998. Differentiation of naive CTL to effector and memory CTL: correlation of effector function with phenotype and cell division. J Immunol 161:5338–5346. [PubMed] [Google Scholar]
- 51.Lin EH, Salon C, Brambilla E, Lavillette D, Szecsi J, Cosset FL, Coll JL. 2010. Fusogenic membrane glycoproteins induce syncytium formation and death in vitro and in vivo: a potential therapy agent for lung cancer. Cancer Gene Ther 17:256–265. doi: 10.1038/cgt.2009.74. [DOI] [PubMed] [Google Scholar]
- 52.Bateman AR, Harrington KJ, Kottke T, Ahmed A, Melcher AA, Gough MJ, Linardakis E, Riddle D, Dietz A, Lohse CM, Strome S, Peterson T, Simari R, Vile RG. 2002. Viral fusogenic membrane glycoproteins kill solid tumor cells by nonapoptotic mechanisms that promote cross presentation of tumor antigens by dendritic cells. Cancer Res 62:6566–6578. [PubMed] [Google Scholar]
- 53.Marsac D, Loirat D, Petit C, Schwartz O, Michel ML. 2002. Enhanced presentation of major histocompatibility complex class I-restricted human immunodeficiency virus type 1 (HIV-1) Gag-specific epitopes after DNA immunization with vectors coding for vesicular stomatitis virus glycoprotein-pseudotyped HIV-1 Gag particles. J Virol 76:7544–7553. doi: 10.1128/JVI.76.15.7544-7553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mao CP, Hung CF, Kang TH, He L, Tsai YC, Wu CY, Wu TC. 2010. Combined administration with DNA encoding vesicular stomatitis virus G protein enhances DNA vaccine potency. J Virol 84:2331–2339. doi: 10.1128/JVI.01954-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nitta S, Sakamoto N, Nakagawa M, Kakinuma S, Mishima K, Kusano-Kitazume A, Kiyohashi K, Murakawa M, Nishimura-Sakurai Y, Azuma S, Tasaka-Fujita M, Asahina Y, Yoneyama M, Fujita T, Watanabe M. 2013. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 57:46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- 56.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol 69:1575–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin C, Pragai BM, Grakoui A, Xu J, Rice CM. 1994. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol 68:8147–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin C, Wu JW, Hsiao K, Su MS. 1997. The hepatitis C virus NS4A protein: interactions with the NS4B and NS5A proteins. J Virol 71:6465–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nace G, Evankovich J, Eid R, Tsung A. 2012. Dendritic cells and damage-associated molecular patterns: endogenous danger signals linking innate and adaptive immunity. J Innate Immun 4:6–15. doi: 10.1159/000334245. [DOI] [PubMed] [Google Scholar]
- 60.Joffre OP, Segura E, Savina A, Amigorena S. 2012. Cross-presentation by dendritic cells. Nat Rev Immunol 12:557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- 61.Steinman RM. 2012. Decisions about dendritic cells: past, present, and future. Annu Rev Immunol 30:1–22. doi: 10.1146/annurev-immunol-100311-102839. [DOI] [PubMed] [Google Scholar]
- 62.Joffre O, Nolte MA, Sporri R, Reis e Sousa C. 2009. Inflammatory signals in dendritic cell activation and the induction of adaptive immunity. Immunol Rev 227:234–247. doi: 10.1111/j.1600-065X.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- 63.Trumpfheller C, Longhi MP, Caskey M, Idoyaga J, Bozzacco L, Keler T, Schlesinger SJ, Steinman RM. 2012. Dendritic cell-targeted protein vaccines: a novel approach to induce T-cell immunity. J Intern Med 271:183–192. doi: 10.1111/j.1365-2796.2011.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frelin L, Alheim M, Chen A, Soderholm J, Rozell B, Barnfield C, Liljestrom P, Sallberg M. 2003. Low dose and gene gun immunization with a hepatitis C virus nonstructural (NS) 3 DNA-based vaccine containing NS4A inhibit NS3/4A-expressing tumors in vivo. Gene Ther 10:686–699. doi: 10.1038/sj.gt.3301933. [DOI] [PubMed] [Google Scholar]
- 65.Seder RA, Darrah PA, Roederer M. 2008. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol 8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 66.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lichterfeld M, Yu XG, Waring MT, Mui SK, Johnston MN, Cohen D, Addo MM, Zaunders J, Alter G, Pae E, Strick D, Allen TM, Rosenberg ES, Walker BD, Altfeld M. 2004. HIV-1-specific cytotoxicity is preferentially mediated by a subset of CD8+ T cells producing both interferon-gamma and tumor necrosis factor-alpha. Blood 104:487–494. doi: 10.1182/blood-2003-12-4341. [DOI] [PubMed] [Google Scholar]
- 68.Bartenschlager R, Lohmann V, Wilkinson T, Koch JO. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J Virol 69:7519–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Basta S, Stoessel R, Basler M, van den Broek M, Groettrup M. 2005. Cross-presentation of the long-lived lymphocytic choriomeningitis virus nucleoprotein does not require neosynthesis and is enhanced via heat shock proteins. J Immunol 175:796–805. doi: 10.4049/jimmunol.175.2.796. [DOI] [PubMed] [Google Scholar]
- 70.Norbury CC, Basta S, Donohue KB, Tscharke DC, Princiotta MF, Berglund P, Gibbs J, Bennink JR, Yewdell JW. 2004. CD8+ T cell cross-priming via transfer of proteasome substrates. Science 304:1318–1321. doi: 10.1126/science.1096378. [DOI] [PubMed] [Google Scholar]
- 71.Shen L, Rock KL. 2004. Cellular protein is the source of cross-priming antigen in vivo. Proc Natl Acad Sci U S A 101:3035–3040. doi: 10.1073/pnas.0308345101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolkers MC, Brouwenstijn N, Bakker AH, Toebes M, Schumacher TN. 2004. Antigen bias in T cell cross-priming. Science 304:1314–1317. doi: 10.1126/science.1096268. [DOI] [PubMed] [Google Scholar]