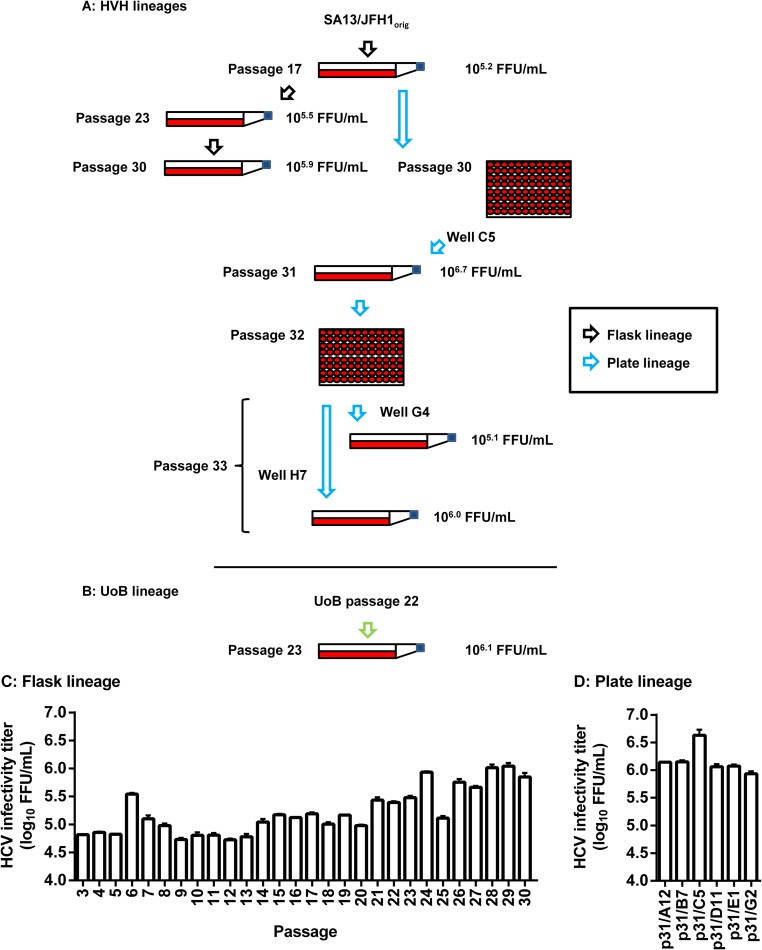
FIG 1.
Flow chart and HCV infectivity titers of serial passage lineages of a genotype 5a Core-NS2 recombinant. (A) A previously described second-passage SA13/JFH1orig virus (18) was serially passaged in cell culture flasks for a total of 30 passages as described in Materials and Methods (black arrows). From the 17th passage, the plate lineage was generated (blue arrows). In the plate lineage, virus was passaged in 96-well plates as described in Materials and Methods until passage 30. From the passage 30 plate, six individual wells were used to infect Huh7.5 cultures to generate passage 31 stocks. SA13/JFH1p31/C5 was subjected to endpoint titration in a 96-well plate (passage 32) in order to obtain individual quasispecies. Six individual wells were selected at the highest possible dilution (endpoint) and used to infect Huh7.5 cultures to generate passage 33 stocks. Representative stocks are shown. (B) Simultaneously, an independently serially passaged passage 22 SA13/JFH1orig stock was received from the University of Birmingham (United Kingdom) (UoB lineage, green arrow). This virus stock was used to generate a passage 23 virus stock (SA13/JFH1p23/UoB). (A and B) Selected serially passaged virus stocks from all lineages are shown. The ORFs of RNA genomes recovered from these stocks were all sequenced (Table 1). Furthermore, the infectivities of the viruses were titrated (Table 1 and data not shown); titers are given as focus-forming units per milliliter. (C) Flask lineage peak infectivity titers of passages 3 to 30. (D) Plate lineage peak infectivity titers. Cultures were generated by infection with supernatant from selected wells from the passage 30 plate. Passage cultures are named according to passage number (p31) and the number of the well from which supernatant for infection was recovered (A12, B7, C5, D11, E1, and G2). (C and D) Supernatant infectivity titers are shown as means (focus-forming units per milliliter) from three replicates with SEM.