Figure 4. Whole-Exome Sequencing of 1 Patient's Primary and Metastatic Tumors.
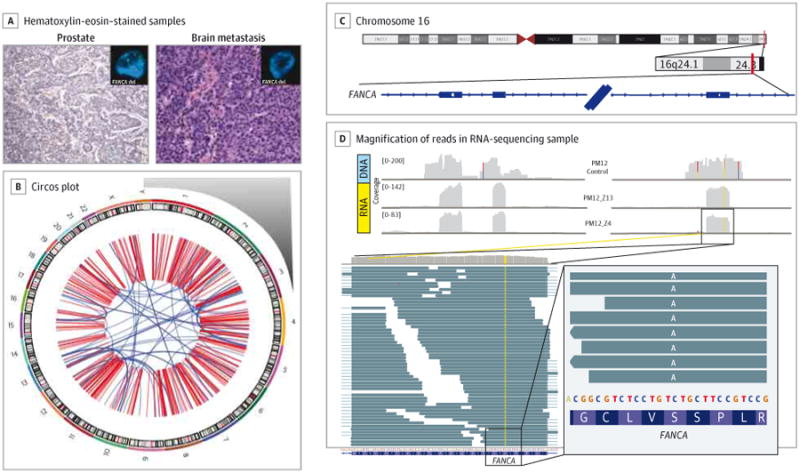
A, Both primary tumor (prostate, histologic subtype Gleason 9 adenocarcinoma with focal neuroendocrine differentiation) and metastatic tumor (brain, histologic subtype neuroendocrine prostate cancer) of patient PM12 were sequenced by means of whole-exome sequencing (WES). A substantial number of somatic mutations and copy number alterations were observed by both WES and whole-genome sequencing. Images are original magnification ×200. B, The whole-genome sequencing Circos plot of PM12's metastatic tumor illustrated a highly altered genome with complex structural variations and rearrangements. Chromosome number is indicated outside the circle. One important finding from WES was hemizygous deletion of the DNA repair gene FANCA in both primary tumor and metastasis, which was confirmed by fluorescence in situ hybridization (tumor panel insets; green probe = control centromeric; red probe = FANCA). C and D, Loss of heterozygosity in FANCA. In the tumor cells, in the heterozygous germline single-nucleotide polymorphism on exon 33 of FANCA (rs17233497), only the missense variant is expressed. C, Coverage of the region for germline (DNA) and 2 tumor samples (RNA). D, Magnification of the reads in 1 RNA-sequencing sample. Reads are reported as on the forward strand; hence, the A variant is indicated, corresponding to a T in the mRNA molecule because FANCA is transcribed from the reverse strand.
