Figure 1. Stimulus-induced translocation of RNase III enzymes.
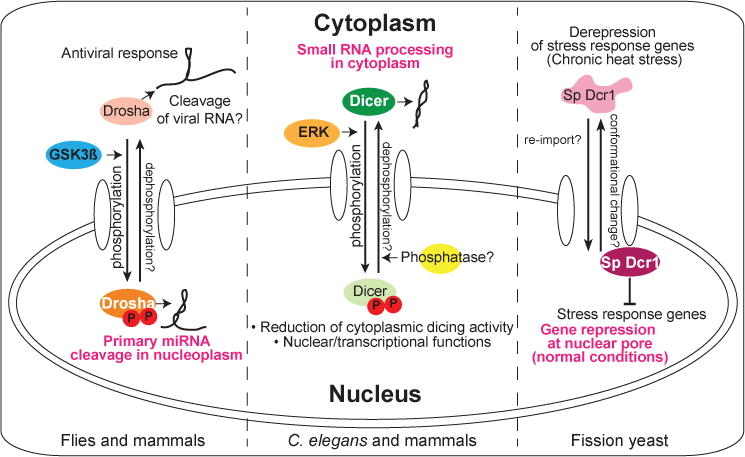
The primary locations of RNase III enzymes under normal conditions or in most cell types are designated by white lettering. (Left) Drosha is normally phosphorylated (by GSK3ß) for nuclear localization and cleaves primary miRNA. Viral infection induces its cytoplasmic accumulation, where it has antiviral activity. (Middle) Dicer is generally cytoplasmically localized to process small RNA precursors. During C. elegans oogenesis or in FGF-stimulated mammalian cells, activated MAPK/ERK phosphorylates Dicer for nuclear translocation, leading to reduced dicing activity in the cytoplasm. Nuclear functions of Dicer have also been proposed. (Right) In fission yeast, stress response genes are suppressed by Dicer (Sp Dcr1) via cotranscriptional silencing at nuclear pores. Chronic heat shock triggers Dcr1 translocation to the cytoplasm, resulting in gene derepression. Current studies of these examples have focused on “one-way” trafficking, but in principle, all of these translocations might be reversible (as designated by question marks).
