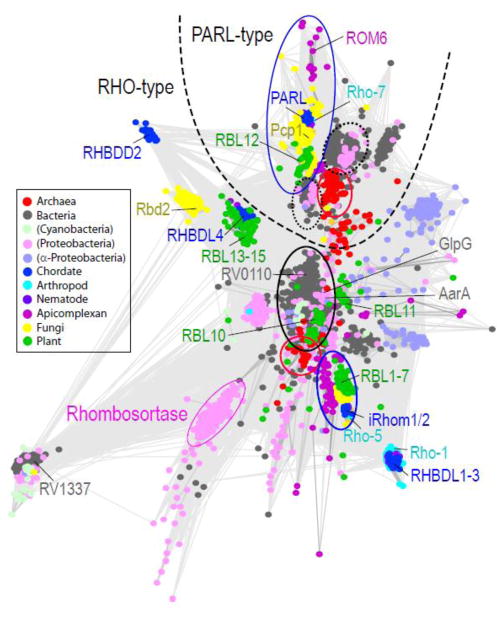
Figure 1. Network-based rhomboid protease clusters.
Network-based CLANS [47] clustering of rhomboids defined by PFAM [48]. Divergent sequences such as derlins were excluded and bacterial representatives were filtered at 80% identity. The resulting 3345 nodes represent individual rhomboid sequences. Nodes are connected by lines that stand for pairwise BLAST E-values (cutoff 1e-7) and are colored according to taxonomy: chordate (blue), arthropod (cyan), nematode (purple), apicomplexan (magenta), fungi (yellow), plant (green), archaea (red), cyanobacteria (light green), proteobacteria (pink), alpha-proteobacteria (slate), and all other bacteria (gray); key rhomboid protease representatives are labeled to the side of their clusters and colored as above. A dashed line separates two major rhomboid protease groupings into PARL-type and RHO-type. Each broad group includes central clusters that segregate according to taxonomy: bacteria circled in black or dotted black, archaea circled in red, and eukaryota circled in blue. Sequences that fall within these centralized groups are listed in the Supplemental table.