Figure 3. Rhomboid protease structure and mechanism suggests convergence of serine protease activity.
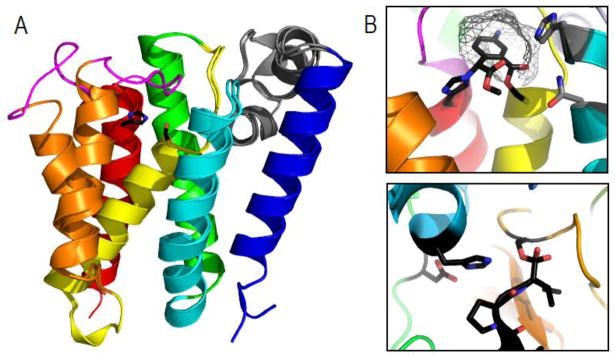
A) GlpG 6TMH transmembrane rhomboid protease core (TMHs colored in rainbow) forms open (PDB ID: 2nrf_A) and closed (PDB ID: 2xov) states dictated by movement of TMH5 (orange) and the loop5 cap (magenta) that covers the active site containing a serine-histidine catalytic dyad (black sticks). An unusual helical loop L1 (gray) is partially submerged in the membrane bilayer. B) Upper panel illustrates a zoom of the GlpG active site (PDB ID: 2oxw) covalently modified by an inhibitor (black) that highlights proximity of catalytic dyad (black stick) to residues that may assist oxyanion formation during catalysis (gray sticks), the presumed S1 pocket (gray wireframe), and the relative orientation of the serine nucleophile with respect to the inhibitor suggesting a si-face attack. C) A zoom of a chymotrypsin active site (PDB ID: 1haz) covalently modified by an inhibitor (black) highlights the catalytic triad (black sticks) with its nucleophilic serine on the opposite face of the inhibitor.
