Figure 4.
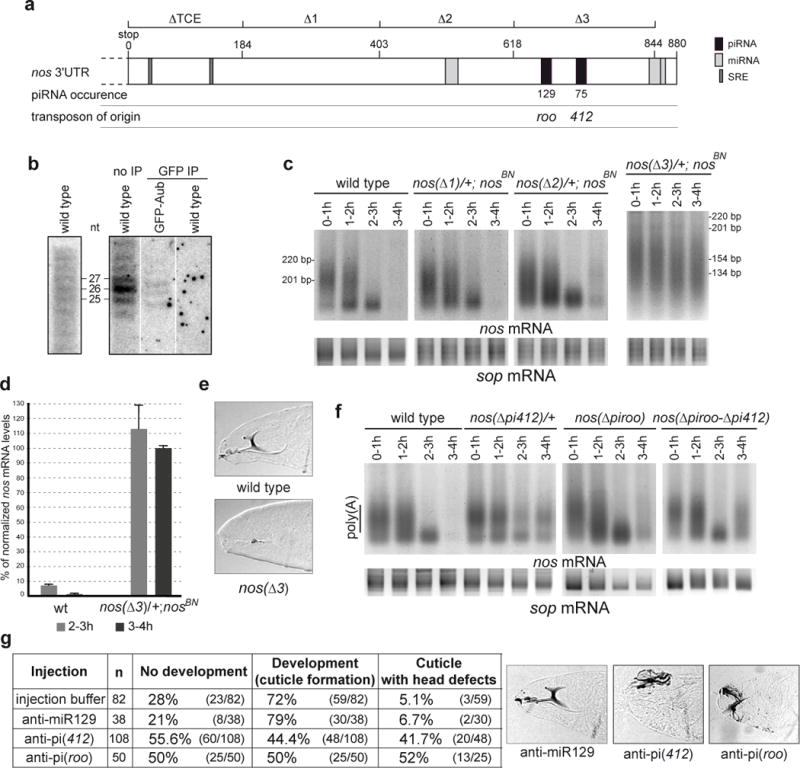
piRNAs target a specific region in nos 3′-UTR which is required for nos mRNA deadenylation. (a) Schematic representation of nos 3′-UTR. The regions deleted in nos genomic transgenes (nos(Δ)) are indicated on the upper line16. SRE, piRNA and miRNA target sites are indicated. Predictions of miRNA targeted regions are from miRBase (miR-31a, miR-314 and miR-263b from proximal to distal). piRNA occurrences in the data sets19,20 are indicated. (b) Northern blots of 0–2 hour embryos probed with riboprobes corresponding to sense nos 3′-UTR (position 403–844) (left panel) and to antisense 412 piRNA (right panel). Anti-GFP immunoprecipitations (GFP IP) were performed using wild-type and GFP-Aub expressing embryos. (c) nos PAT assays. For nos(Δ3) the fragment amplified in the PAT assay is shorter than the fragment amplified in the other nos PAT assays (Supplementary Figure 9). (d) Quantification of nos mRNA levels from the nos(Δ3) transgene by RT-QPCR. Mean value of three quantifications, error bars correspond to s.d. (e) Cuticle preparations of embryos from nos(Δ3) females (lack of head skeleton). (f) PAT assays of embryos with nos genomic transgenes in which sequences complementary to 412 piRNA, roo piRNA, or both sequences have been deleted. The sop mRNA was used as a control in c and f. (g) Injection of 2′ O-methyl anti-piRNA in embryos. Control injections were with injection buffer alone or with the irrelevant anti-miR129. Examples of cuticles following injections of anti-miR129 (wild-type head skeleton), anti-pi(412) and anti-pi(roo) (affected head skeleton).
