Abstract
Campylobacters are a major global health burden and a cause of food-borne diarrheal illness and economic loss worldwide. In developing countries, Campylobacter infections are frequent in children under age two and may be associated with mortality. In developed countries, they are a common cause of bacterial diarrhea in early adulthood. In the United States, antibiotic resistance against Campylobacter is notably increased from 13% in 1997 to nearly 25% in 2011. Novel drug targets are urgently needed but remain a daunting task to accomplish. We suggest that omics-guided drug discovery is timely and worth considering in this context. The present study employed an integrated subtractive genomics and comparative metabolic pathway analysis approach. We identified 16 unique pathways from Campylobacter when compared against H. sapiens with 326 non-redundant proteins; 115 of these were found to be essential in the Database of Essential Genes. Sixty-six proteins among these were non-homologous to the human proteome. Six membrane proteins, of which four are transporters, have been proposed as potential vaccine candidates. Screening of 66 essential non-homologous proteins against DrugBank resulted in identification of 34 proteins with drug-ability potential, many of which play critical roles in bacterial growth and survival. Out of these, eight proteins had approved drug targets available in DrugBank, the majority serving crucial roles in cell wall synthesis and energy metabolism and therefore having the potential to be utilized as drug targets. We conclude by underscoring that screening against these proteins with inhibitors may aid in future discovery of novel therapeutics against campylobacteriosis in ways that will be pathogen specific, and thus have minimal toxic effect on host. Omics-guided drug discovery and bioinformatics analyses offer the broad potential for veritable advances in global health relevant novel therapeutics.
Introduction
Campylobacters are a major cause of food-borne diarrheal illness and result in a high morbidity and mortality rate, and economic loss in every region of the world (WHO, 2011). In developing countries, Campylobacter infections are frequent in children under age 2, sometimes leading to death. In industrialized nations, they are most frequently identified cause of bacterial diarrhea in early adulthood (CDC, 2000). According to a report released by Centers for Disease Control and Prevention (CDC), there are 1.3 million incidences of campylobacteriosis and there is rapid escalation of antibiotic resistance in Campylobacter from 13% in 1997 to almost 25% in 2011 in United States (CDC, 2013). The growing body of literature has documented that resistance to antibiotics such as quinolones, macrolides, tetracyclines, chloramphenicol, cephalosporins, and aminoglycosides is increasing rapidly in most parts of the world due to common and indiscriminate use of these agents (Akhtar, 1988; Engberg et al., 2001; Hoge et al., 1998; Reina et al., 1994). Campylobacters are highly important from a socioeconomic perspective, which strongly indicates a need for novel therapeutic targets with a high potential to improve quality of life and survival rates.
Since the publication of pathogenic bacterial genome sequences Haemophilus influenzae (Fleischmann et al., 1995) and Mycoplasma genetalium (Fraser et al., 1995), in 1995, the number of completed genome sequence for various microbial species has increased rapidly. These data in the post-genomic era have provided researchers with the possibility to exploit it fully for identification of novel therapeutic targets and have opened up new avenues for genome-wide application of comparative and subtractive genomics approaches for therapeutic intervention.
The subtractive genomics approach has been advertently used by many researchers (Chawley et al., 2014; Ghosh et al., 2014; Samal et al., 2015; Sarangi et al., 2009) in search of novel drug targets for various microbes such as Vibrio cholerae, Staphylococcus aureus, Salmonella typhimurium, and Neisseria meningitides, respectively. Genome sequences of several Campylobacter species have been published including Campylobacter concisus, C. curvus, C. fetus, C. hominis, and six strains of C. jejuni (http://gcid.jcvi.org/projects/msc/campylobacter/). In this study we report the subtractive genomics approach integrated with comparative metabolic pathway analysis aimed at identifying novel therapeutic target proteins of C. jejuni pathogenic strain NCTC11168.
Methods
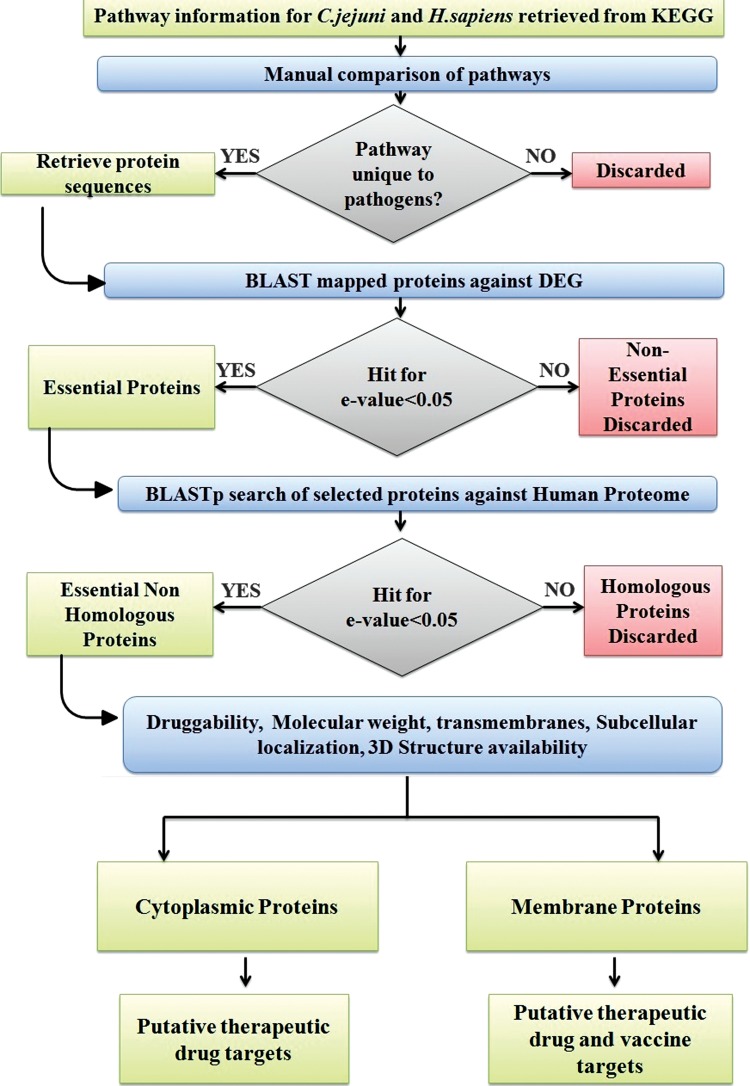
Various databases and tools as described in the workflow (Fig. 1) were utilized for the identification of putative therapeutic targets against C. jejuni, integrating a subtractive genomics approach with genome-wide comparative pathway analysis.
Fig. 1.
Schematic workflow of comparative genomics approach to identify drug targets in pathogenic C.jejuni strain NCTC11168.
Host and pathogen comparative metabolic pathway analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000), the most comprehensive resource of pathway information, was used for comparative genome-wide pathway analysis of C. jejuni and Homo sapiens. Manual comparison was done to identify the pathways unique to C. jejuni as per KEGG database annotations. Protein sequences for the enzymes involved in the unique pathways were retrieved from the Uniprot protein database.
Subtractive genomics and identification of essential non-homologous pathogen proteins
Essential non-homologous proteins from pathogen proteome were selected by a two-step comparison. In the first comparison, a subtractive genomics approach was applied to pathogen proteins from unique pathways. The BLAST search (Altschul et al., 1990) based on essentiality criteria was performed against the Database of Essential Genes (DEG version 10.9) (Zhang et al., 2004), which hosts records of essential genomic elements critical for an organism's survival, such as protein-coding genes and non-coding RNAs, among bacteria, archaea, and eukaryotes. An expectation value (e-value) threshold of 0.005 was used as filtering criteria for BLAST hits; with C. jejuni and Helicobacter pylori as background organisms against which a similarity search for identification of essential genes was performed. In the second step comparison, essential pathogen proteins were further screened on the basis of homology with the host proteome at an e-value cut off of 0.05. In the BLASTP search, proteins that did not have any hits below e-value inclusion threshold were selected to be essential non-homologous proteins from C. jejuni.
Prioritization of essential non-homologous proteins for therapeutic targets
The molecular and structural properties of essential non-homologous pathogen proteins were calculated for aid in prioritization of the drug targets that will most likely lead to effective treatments. Subcellular localization of identified proteins was predicted by PSORTb (Yu et al., 2010), which uses a Bayesian network model to calculate associated probability value for five major localization sites viz. cytoplasmic, inner membrane, periplasmic, outer membrane, and extracellular with p-value criteria 7.5. TMHMM server, which is a Hidden Markov Model (HMM)-based tool for prediction of alpha helices in membrane proteins, was used for transmembrane predictions (Krogh et al., 2001).
A search was performed to identify the proteins for which experimentally or computationally solved structures are available in PDB (Berman et al., 2000), Modbase (Pieper et al., 2006), or Protein Model Portal (Haas et al., 2013). A search of identified proteins in Uniprot was performed for retrieving information pertaining to molecular weight and existence of proteins. The information on protein existence provides different types of evidence for experimental characterization at protein and/or transcript level, homology inference, or uncertainty.
The VaxiJen server (Doytchinova and Flower, 2007) was employed to check the antigenicity of the membrane-localized proteins with a threshold of 0.6. The antigenic sequences were further screened for their ability to bind to MHC Class I molecule using ProPred-I (Singh and Raghava, 2003). ProPred-I implements proteasomal processing with matrices for 47 MHC Class-I alleles to identify the regions in the antigenic sequences that can act as potential MHC binders.
Drug-ability of essential son-homologous proteins
To evaluate the drug-ability potential, which is the ability of a particular biological target to bind with high affinity to known drugs of each identified therapeutic target, we subjected each protein to a BLASTP search against DrugBank with an e-value 0.01. DrugBank is a unique comprehensive resource that integrates drug data with drug target information at sequence, structure, and pathway levels (Wishart et al., 2008). DrugBank version 4.2 currently has information about 7737 drug entries that include 1585 FDA-approved small molecule drugs, 158 FDA-approved biotech (protein/peptide) drugs, 89 nutraceuticals, and over 6000 experimental drugs, along with 4281 non-redundant protein sequences (drug target/enzyme/transporter/carrier) linked to these drugs.
Results
Identification of unique metabolic pathways
The KEGG database initiated in 1995 is a computational representation for biological systems, integrating genetic information of genes and proteins with chemical and systemic information of molecular interaction and reaction networks. It links genetic building block information with higher order functional information. Currently KEGG database houses 87 different pathways for C. jejuni 11168 strain and 292 pathways for H. sapiens. As described in the workflow (Fig. 1), a manual comparison of host and pathogen pathways resulted in identification of 16 unique pathways to the pathogen (Table 1), with 71 remaining pathways being shared by both humans and Campylobacters. Furthermore, the proteins involved in unique pathways were identified.
Table 1.
Metabolic Pathways Unique to C. jejuni Against Human Host and Proteins Associated with Corresponding Pathway
| No. | Pathway name | Pathway ID | No. of proteins |
|---|---|---|---|
| 1 | Carbapenem biosynthesis | cje0033 | 2 |
| 2 | Novobiocin biosynthesis | cje00401 | 4 |
| 3 | D-Alanine metabolism | cje00473 | 2 |
| 4 | Streptomycin biosynthesis | cje00521 | 3 |
| 5 | Lipopolysaccharide biosynthesis | cje00540 | 20 |
| 6 | Peptidoglycan biosynthesis | cje00550 | 14 |
| 7 | C5-Branched dibasic acid metabolism | cje00660 | 7 |
| 8 | Methane metabolism | cje00680 | 15 |
| 9 | Biosynthesis of secondary metabolites | cje01110 | 167 |
| 10 | Microbial metabolism in diverse environments | cje01120 | 88 |
| 11 | beta-Lactam resistance | cje01501 | 5 |
| 12 | Vancomycin resistance | cje01502 | 5 |
| 13 | Two-component system | cje02020 | 43 |
| 14 | Bacterial chemotaxis | cje02030 | 20 |
| 15 | Flagellar assembly | cje02040 | 33 |
| 16 | Bacterial secretion system | cje03070 | 18 |
Identification of essential non-homologous pathogen proteins
A total of 446 proteins were identified to be involved in 16 unique pathways. Few proteins were involved in more than one pathway, which resulted in 326 protein sequences from unique pathways after removing redundant protein sequences. A BLASTP search of these 326 protein sequences against 551 essential genes from C. jejuni and H. pylori in DEG revealed a total of 115 essential protein sequences with hits below e-value 0.05.
These essential protein sequences were further filtered out by homology search against human proteome for identification of non-homologous protein sequences. This comparison detected 66 essential non-homologous proteins with no hits against H. sapiens below e-value 0.05. This comparison was performed to identify proteins unique to pathogen so as to avoid adverse effects on the human host (Butt et al., 2012), as the potential drug may also target host enzymes. These 66 essential non-homologous protein sequences represent the potential to be further exploited for therapeutic drug design against C. jejuni.
Prioritization of essential non-homologous proteins for therapeutic drug targets
Although all 66 essential non-homologous identified proteins are potential drug targets, these can be further filtered using additional prioritization parameters. Subcellular localization prediction using PSORTb program identified 63 proteins to be of cytoplasmic origin and all the essential non-homologous proteins were found to be <110 kDa. A search for availability of 3D structure identified seven proteins for which there was no structure available, while for six of these proteins experimentally solved structures were available. The remaining 53 proteins had computationally solved structures available in either Modbase or ProteinModel Portal. TMHHM server predicted 16 proteins that had one or more helices traversing the membrane, six of which were found to be antigenic above the specified threshold. ProPred-I server predicted MHC binder regions in all of the six proteins (murF, frdC, ccoP, secD, Cj1094c, and tatC) for different MHC class I alleles, and these proteins represent potential vaccine candidates as most of them are transporters, surface exposed proteins. All these results are presented in Table 2.
Table 2.
Essential Non-Homologous Proteins of C. jejuni with Results of Prioritization Analysis (Transmembrane and Cellular Location Prediction)
| No. | Gene name | Protein Id | Pathway involved | Protein existence | Subcellular location | TMHMM | 3D Exp | 3D Model | MW (Da) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | alr | Q9PP26 | D-Alanine metabolism; Vancomycin resistance | Inferred | Cytoplasmic | No | No | Yes | 37,256 |
| 2 | ddl | Q9PPC2 | D-Alanine metabolism; Vancomycin resistance; Peptidoglycan biosynthesis | Inferred | Cytoplasmic | No | No | Yes | 39,908 |
| 3 | lpxA | Q9PIM1 | Lipopolysaccharide biosynthesis | Predicted | Cytoplasmic | No | Yes | Yes | 28,651 |
| 4 | lpxD | Q9PHU0 | Lipopolysaccharide biosynthesis | Inferred | Unknown | No | No | Yes | 34,679 |
| 5 | lpxC | Q9PIZ5 | Lipopolysaccharide biosynthesis | Inferred | Cytoplasmic | No | No | Yes | 32,883 |
| 6 | murA | Q9PP65 | Peptidoglycan biosynthesis | Inferred | Cytoplasmic | No | No | Yes | 45,186 |
| 7 | murB | Q9PM01 | Peptidoglycan biosynthesis | Inferred | Cytoplasmic | No | No | Yes | 28,994 |
| 8 | murC | Q9PNN7 | Peptidoglycan biosynthesis | Inferred | Cytoplasmic | No | No | Yes | 48,372 |
| 9 | murF | Q0PA95 | Peptidoglycan biosynthesis; Vancomycin resistance | Predicted | Cytoplasmic Membrane | Yes | No | Yes | 55,234 |
| 10 | pbpA | Q0PB07 | Peptidoglycan biosynthesis; Beta-Lactam resistance | Predicted | Cytoplasmic Membrane | Yes | No | Yes | 72,628 |
| 11 | fba | Q0PAS0 | Methane metabolism; Biosynthesis of secondary metabolites; Microbial metabolism in diverse environments | Predicted | Cytoplasmic | No | Yes | Yes | 38,730 |
| 12 | aroQ | Q9PJ53 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | No | Yes | 17,595 |
| 13 | pfs | Q0PC20 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | No | Yes | 25,225 |
| 14 | panD | Q9PIK3 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | Yes | Yes | 13,974 |
| 15 | panC | Q9PIK2 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | Yes | Yes | 32,087 |
| 16 | panB | Q9PIK1 | Biosynthesis of secondary metabolites | Inferred | Cytoplasmic | No | No | Yes | 30,202 |
| 17 | pheA | Q0PBJ3 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | No | Yes | 40,477 |
| 18 | aroE | Q0PBA5 | Biosynthesis of secondary metabolites | Inferred | Cytoplasmic | No | No | Yes | 29,978 |
| 19 | trpC | Q9PI11 | Biosynthesis of secondary metabolites | Inferred | Cytoplasmic | No | No | Yes | 29,573 |
| 20 | ispH | P0C632 | Biosynthesis of secondary metabolites | Inferred | Cytoplasmic | No | No | No | 31,551 |
| 21 | aroA | P0C630 | Biosynthesis of secondary metabolites | Inferred | Cytoplasmic | No | No | Yes | 47,288 |
| 22 | aroB | Q9PNT2 | Biosynthesis of secondary metabolites | Inferred | Cytoplasmic | No | No | Yes | 39,596 |
| 23 | dxr | Q9PMV3 | Biosynthesis of secondary metabolites | Inferred | Cytoplasmic | No | No | Yes | 39,548 |
| 24 | Cj1370 | Q0P8N9 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | No | Yes | 17,035 |
| 25 | aroC | Q9PM41 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | Yes | Yes | 39,252 |
| 26 | dapB | Q9PIT2 | Biosynthesis of secondary metabolites; Microbial metabolism in diverse environments | Inferred | Cytoplasmic | No | No | Yes | 26,696 |
| 27 | rpiB | Q0P9X4 | Biosynthesis of secondary metabolites; Microbial metabolism in diverse environments | Predicted | Cytoplasmic | No | No | Yes | 16,293 |
| 28 | pbpB | Q0PAZ1 | Peptidoglycan biosynthesis; Beta-Lactam resistance | Predicted | Cytoplasmic Membrane | Yes | No | Yes | 68,378 |
| 29 | pbpC | Q0PAL6 | Peptidoglycan biosynthesis; Beta-Lactam resistance | Predicted | Cytoplasmic Membrane | Yes | No | Yes | 67,971 |
| 30 | frdC | Q0PBA2 | Biosynthesis of secondary metabolites; Microbial metabolism in diverse environments; Two-component system | Predicted | Cytoplasmic Membrane | Yes | No | Yes | 30,028 |
| 31 | ccoP | Q0P8D0 | Two-component system | Inferred | Periplasmic | Yes | No | Yes | 31,162 |
| 32 | cheA | Q0PBM2 | Two-component system; Bacterial chemotaxis | Predicted | Cytoplasmic | No | No | Yes | 85,300 |
| 33 | cheV | Q0PBM1 | Two-component system; Bacterial chemotaxis | Predicted | Cytoplasmic | No | No | Yes | 35,858 |
| 34 | cheY | P0C635 | Two-component system; Bacterial chemotaxis | Inferred | Cytoplasmic | No | No | Yes | 14,437 |
| 35 | kdtA | Q0PAH2 | Lipopolysaccharide biosynthesis | Predicted | Cytoplasmic | Yes | No | Yes | 45,217 |
| 36 | gmhB | Q6TG07 | Lipopolysaccharide biosynthesis | Inferred | Cytoplasmic | No | No | Yes | 20,246 |
| 37 | waaC | Q0P9C1 | Lipopolysaccharide biosynthesis | Predicted | Cytoplasmic | No | No | Yes | 39,350 |
| 38 | Cj0462 | Q0PB51 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | No | Yes | 39,759 |
| 39 | ubiD | Q0PAX0 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | No | Yes | 69,160 |
| 40 | ispG | Q9PPM1 | Biosynthesis of secondary metabolites | Inferred | Cytoplasmic | No | No | Yes | 39,366 |
| 41 | Cj0837c | Q0PA53 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | No | No | 36,994 |
| 42 | hisH1 | Q0P8U2 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | No | Yes | 22,699 |
| 43 | Cj1368 | Q0P8P1 | Biosynthesis of secondary metabolites | Predicted | Cytoplasmic | No | No | Yes | 41,278 |
| 44 | hisB | Q9PM76 | Biosynthesis of secondary metabolites | Inferred | Cytoplasmic | No | No | Yes | 39,580 |
| 45 | oorD | Q0PAY1 | Microbial metabolism in diverse environments | Predicted | Unknown | No | No | Yes | 11,443 |
| 46 | oorA | Q0PAY0 | Microbial metabolism in diverse environments | Predicted | Cytoplasmic | No | No | Yes | 41,052 |
| 47 | oorB | Q0PAX9 | Microbial metabolism in diverse environments | Predicted | Cytoplasmic | No | No | Yes | 31,208 |
| 48 | oorC | Q0PAX8 | Microbial metabolism in diverse environments | Predicted | Cytoplasmic | No | Yes | Yes | 20,097 |
| 49 | lysC | Q0PAT5 | Biosynthesis of secondary metabolites; Microbial metabolism in diverse environments | Inferred | Unknown | No | No | Yes | 42,676 |
| 50 | csrA | Q0P9F1 | Two-component system | Predicted | Unknown | No | No | Yes | 8,441 |
| 51 | Cj1153 | Q0P9A3 | Microbial metabolism in diverse environments; Two-component system | Predicted | Periplasmic | Yes | No | Yes | 10,838 |
| 52 | ccoN | Q0P8C7 | Two-component system | Inferred | Cytoplasmic Membrane | Yes | No | Yes | 55,888 |
| 53 | ccoQ | Q0P8C9 | Two-component system | Predicted | Unknown | Yes | No | No | 10,376 |
| 54 | fliE | Q9PHY8 | Flagellar assembly | Inferred | Unknown | No | No | No | 10,830 |
| 55 | flgA | Q0PAC1 | Flagellar assembly | Predicted | Cytoplasmic | No | No | Yes | 25,114 |
| 56 | flaD | Q0PA11 | Flagellar assembly | Predicted | Extracellular | No | No | Yes | 82,051 |
| 57 | flaB | P56964 | Two-component system; Flagellar assembly | Inferred | Extracellular | No | No | Yes | 59,186 |
| 58 | flaA | P56963 | Two-component system; Flagellar assembly | Inferred | Extracellular | No | No | Yes | 59,039 |
| 59 | flaC | P96747 | Two-component system; Flagellar assembly | Predicted | Periplasmic | No | No | Yes | 26,606 |
| 60 | secD | Q0P9G1 | Bacterial secretion system | Inferred | Cytoplasmic Membrane | Yes | No | Yes | 57,251 |
| 61 | secF | Q0P9G2 | Bacterial secretion system | Inferred | Cytoplasmic Membrane | Yes | No | Yes | 36,112 |
| 62 | secE | Q0PB41 | Bacterial secretion system | Inferred | Cytoplasmic Membrane | Yes | No | No | 6,745 |
| 63 | secY | Q0P7U3 | Bacterial secretion system | Inferred | Cytoplasmic Membrane | Yes | No | Yes | 46,050 |
| 64 | Cj1094c | Q0P9G0 | Bacterial secretion system | Predicted | Cytoplasmic Membrane | Yes | No | Yes | 10,172 |
| 65 | tatB | Q9PHT7 | Bacterial secretion system | Inferred | Cytoplasmic | No | No | No | 15,746 |
| 66 | tatC | Q9PHT8 | Bacterial secretion system | Inferred | Cytoplasmic Membrane | Yes | No | No | 27,808 |
Drug-ability of potential drug targets
To examine the drug-ability of each of the essential non-homologous proteins, they were subjected to a BLASTP search against all the drug-targeted proteins in DrugBank database, which resulted in the identification of 34 C. jejuni proteins that shared high similarity to the binding partners of these drug-targeted proteins from DrugBank. Nine of these 34 proteins were FDA-approved drugs or nutraceuticals, while the remaining 25 were under experimentation. In Table 3 we have summarized the identified target protein binding partners of the all the drugs of 34 C. jejuni target proteins.
Table 3.
Essential Non-Homologous Proteins of C.jejuni Similar to Binding Partners of Drugs Available in Drug Bank by Homology Search
| No. | Uniprot Id | Gene name | Protein name | Pathway involved | Drug Bank Id | Drug name | Drug group |
|---|---|---|---|---|---|---|---|
| 1 | Q9PP26 | alr | Alanine racemase | D-Alanine metabolism, Vancomycin resistance | DB01993, DB02142, DB03097, DB03327, DB03579, DB03766, DB03801, DB04467, DB03252, DB03801, DB00260 | N-(5′-Phosphopyridoxyl)-D-Alanine, Pyridoxamine-5′-Phosphate, Pmp-Hydroxyisoxazole, Pyridoxamine-5-Phosphate-Hydroxyisoxazole, {1-[(3-Hydroxy-Methyl-5-Phosphonooxy-Methyl-Pyridin-4-Ylmethyl)-Amino]-Ethyl}-Phosphonic Acid, Pyridoxyl-N,O-Cycloserylamide-5-Monophosphate, Propanoic Acid, Lysine Nz-Carboxylic Acid, Pyridoxyl-Alanine-5-Phosphate, D-Lysine, Lysine Nz-Carboxylic Acid, Cycloserine | Experimental (11); Approved |
| 2 | Q9PPC2 | ddl | D-Alanine–D-alanine ligase | D-Alanine metabolism, Vancomycin resistance, Peptidoglycan biosynthesis | DB00260, DB07805 | Cycloserine, 3-chloro-2,2-dimethyl-n-[4-(trifluoromethyl)phenyl]propanamide | Approved; Experimental |
| 3 | Q9PIM1 | lpxA | Acyl-[acyl-carrier-protein]–UDP-N-acetylglucosamine O-acyltransferase | Lipopolysaccharide biosynthesis | DB01694, DB08558 | D-tartaric acid, 2-hydroxymethyl-6-octylsulfanyl-tetrahydro-pyran-3,4,5-triol | Experimental (2) |
| 4 | Q9PHU0 | lpxD | UDP-3-O-acylglucosamine N-acyltransferase | Lipopolysaccharide biosynthesis | DB01694, DB08558 | D-tartaric acid, 2-hydroxymethyl-6-octylsulfanyl-tetrahydro-pyran-3,4,5-triol | Experimental (2) |
| 5 | Q9PIZ5 | lpxC | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase | Lipopolysaccharide biosynthesis | DB07861, DBo1991, DB04257, DB07355, DB07536, DB08231 | (2R)-N-hydroxy-3-naphthalen-2-yl-2-[(naphthalen-2-ylsulfonyl)amino]propanamide, Tu-514, Palmitoleic Acid, 3-(heptyloxy)benzoic acid, N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, myristic acid | Experimental (6) |
| 6 | Q9PP65 | murA | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | Peptidoglycan biosynthesis | DB01879, DB02435, DB02995, DB03089, DB04174, DB04474, DB03397, DB00828, | (S)-2-{Methyl-[2-(Naphthalene-2-Sulfonylamino)-5-(Naphthalene-2-Sulfonyloxy)-Benzoyl]-Amino}-Succinicacid, Aminomethylcyclohexane, Cyclohexylammonium Ion, L-Iso-Aspartate, 3′-1-Carboxy-1-Phosphonooxy-Ethoxy-Uridine-Diphosphate-N-Acetylglucosamine, 1-Anilino-8-Naphthalene Sulfonate, Uridine-Diphosphate-N-Acetylglucosamine, Fosfomycin | Experimental (7); Approved |
| 7 | Q9PM01 | murB | UDP-N-acetylenolpyruvoylglucosamine reductase | Peptidoglycan biosynthesis | DB03147 | Flavin adenine dinucleotide | Approved |
| 8 | Q9PNN7 | murC | UDP-N-acetylmuramate–L-alanine ligase | Peptidoglycan biosynthesis | DB01673, DB03909, DB04395 | Uridine-5′-Diphosphate-N-Acetylmuramoyl-L-Alanine, Adenosine-5′-[Beta, Gamma-Methylene]Triphosphate, Phosphoaminophosphonic Acid-Adenylate Ester | Experimental (3) |
| 9 | Q0PA95 | murF | UDP-N-acetylmuramoyl-tripeptide D-alanyl-D-alanine ligase | Peptidoglycan biosynthesis, Vancomycin resistance | DB06970 | 2-chloro-n-(3-cyano-5,6-dihydro-4h-cyclopenta[b]thiophen-2-yl)-5-diethylsulfamoyl-benzamide | Experimental |
| 10 | Q0PB07 | pbpA | Penicillin-binding protein | Peptidoglycan biosynthesis, beta-Lactam resistance | DB00229, DB00267, DB00301, DB00417, DB00456, DB00713, DB00833, DB00948, DB01000, DB01060, DB01061, DB01066, DB01112, DB01139, DB01147, DB01602, DB01604, DB01605, DB01150, DB00415, DB00438, DB00485, DB00493, DB00567, DB00607, DB01331, DB01603, DB08795, DB00274, DB00430, DB01327, DB01328, DB01329, DB01332, DB01333, DB01414 DB01415, DB01598, DB04570, DB04133, DB03313, DB08375 DB00447, DB01163, DB05659, DB00739, DB01140, DB00303 | Cefotiam, Cefmenoxime, Flucloxacillin, Penicillin V, Cefalotin, Oxacillin, Cefaclor, Mezlocillin, Cyclacillin, Amoxicillin, Azlocillin, Cefditoren, Cefuroxime, Cefapirin, Cloxacillin, Bacampicillin, Pivampicillin, Pivmecillinam, Cefprozil, Ampicillin, Ceftazidime, Dicloxacillin, Cefotaxime, Cephalexin, Nafcillin, Cefoxitin, Meticillin, Azidocillin, Cefmetazole, Cefpiramide, Cefazolin, Cefonicid, Cefoperazone, Ceftizoxime, Cefradine, Cefacetrile, Ceftibuten, Imipenem, Latamoxef, Degraded Cephaloridine, Cephalosporin C, Pcnotaxime group, Loracarbef, Amdinocillin, faropenem medoxomil, Hetacillin, Cefadroxil, Ertapenem | Approved (39); Experimental (3); Withdrawn (2); Investigational; Approved, withdrawn (2); Approved, investigational |
| 11 | Q0PAS0 | fba | Fructose-bisphosphate aldolase | Methane metabolism, Biosynthesis of secondary metabolites, Microbial metabolism in diverse environments | DB03026 | Phosphoglycolohydroxamic Acid | Experimental |
| 12 | Q9PJ53 | aroQ | 3-Dehydroquinate dehydratase | Biosynthesis of secondary metabolites | DB03868, DB04698, DB02801, DB03739, DB02786, DB04347, DB04656, DB08485 | 3-Dehydroquinic Acid, N-(1,4-dihydro-5h-tetrazol-5-ylidene)-9-oxo-9h-xanthene-2-sulfonamide, 2,3 -Anhydro-Quinic Acid, 3-Hydroxyimino Quinic Acid, 2-Anhydro-3-Fluoro-Quinic Acid, 3-Dehydroshikimate, 1,3,4-trihydroxy-5-(3-phenoxypropyl)-cyclohexane-1-carboxylic acid, (1S,4S,5S)-1,4,5-trihydroxy-3-[3-(phenylthio)phenyl]cyclohex-2-ene-1-carboxylic acid | Experimental (8) |
| 13 | Q0PC20 | pfs | Aminodeoxyfutalosine nucleosidase | Biosynthesis of secondary metabolites | DB07463, DB02158, DB02281, DB02933, DB08606, DB07649, DB02857, DB00173 | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, (1s)-1-(9-Deazaadenin-9-Yl)-1,4,5-Trideoxy-1,4-Imino-5-Methylthio-D-Ribitol, Formycin, 5′-Deoxy-5′-(Methylthio)-Tubercidin, 3R,4S)-1-[(4-amino-5h-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(methylsulfanyl)methyl]pyrrolidin-3-ol, (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(benzylsulfanyl)methyl]pyrrolidin-3-ol, Guanosine, Adenine | Experimental (7); Approved, Nutraceutical |
| 14 | Q9PIK3 | panD | Aspartate 1-decarboxylase | Biosynthesis of secondary metabolites | DB02175, DB03382, | Malonic acid, S-Oxy Cysteine | Experimental (2) |
| 15 | Q9PIK2 | panC | Pantothenate synthetase | Biosynthesis of secondary metabolites | DB01930, DB02596, DB02694, DB03107 | 2,4-Dihydroxy-3,3-Dimethyl-Butyrate, Alpha,Beta-Methyleneadenosine-5′-Triphosphate, Pantoyl Adenylate, Beta-Alanine | Experimental (4) |
| 16 | Q9PIK1 | panB | 3-Methyl-2-oxobutanoate hydroxymethyltransferase | Biosynthesis of secondary metabolites | DB03795, DB04074 | 2-Dehydropantoate, Alpha-ketoisovalerate | Experimental (2) |
| 17 | Q0PBJ3 | pheA | Chorismate mutase/prephenate dehydratase | Biosynthesis of secondary metabolites | DB08648, DB03884 | 8-hydroxy-2-oxa-bicyclo[3.3.1]non-6-ene-3,5-dicarboxylic acid, 3-Phenylpyruvic Acid | Experimental (2) |
| 18 | Q0PBA5 | aroE | Shikimate dehydrogenase | Biosynthesis of secondary metabolites | DB02363, DB03461, DB04447 | 2′-Monophosphoadenosine-5′-Diphosphate, 2′-Monophosphoadenosine 5′-Diphosphoribose, 1,4-Dithiothreitol | Experimental (3) |
| 19 | Q9PI11 | trpC | Indole-3-glycerol phosphate synthase | Biosynthesis of secondary metabolites | DB03543 | 1-(O-Carboxy-Phenylamino)-1-Deoxy-D-Ribulose-5-Phosphate | Experimental |
| 20 | P0C632 | ispH | 4-Hydroxy-3-methylbut-2-enyl diphosphate reductase | Biosynthesis of secondary metabolites | DB01785, DB04714 | Dimethylallyl Diphosphate, Isopentenyl Pyrophosphate | Experimental (2) |
| 21 | P0C630 | aroA | 3-Phosphoshikimate 1-carboxyvinyltransferase | Biosynthesis of secondary metabolites | DB04328, DB04539, DB01942, DB03116 | Shikimate-3-Phosphate, Glyphosate, Formic Acid, 5-(1-Carboxy-1-Phosphonooxy-Ethoxyl)-Shikimate-3-Phosphate | Experimental (4) |
| 22 | Q9PNT2 | aroB | 3-Dehydroquinate synthase | Biosynthesis of secondary metabolites | DB02592 | Carbaphosphonate | Experimental |
| 23 | Q9PMV3 | dxr | 1-Deoxy-D-xylulose 5-phosphate reductoisomerase | Biosynthesis of secondary metabolites | DB02496, DB02948, DB03649, DB04272 | 1-Deoxy-D-xylulose 5-phosphate, Fosmidomycin, [{(5-Chloro-2-Pyridinyl)Amino} Methylene]-1,1-Bisphosphonate, Citric Acid | Experimental (3); Nutraceutical |
| 24 | Q0P8N9 | Cj1370 | Biosynthesis of secondary metabolites | DB02075 | (1s)-1(9-Deazahypoxanthin-9yl)1,4-Dideoxy-1,4-Imino-D-Ribitol-5-Phosphate | Experimental | |
| 25 | Q9PM41 | aroC | Chorismate synthase | Biosynthesis of secondary metabolites | DB03247, DB03350, DB03969, DB04267 | Riboflavin Monophosphate, Cobalt Hexammine Ion, 3-Acetyl Pyridine Adenine Dinucleotide, Dipicolinic Acid | Experimental (4) |
| 26 | Q9PIT2 | dapB | 4-Hydroxy-tetrahydrodipicolinate reductase | Biosynthesis of secondary metabolites, Microbial metabolism in diverse environments | DB03969, DB04267 | 3-Acetyl Pyridine Adenine Dinucleotide, Dipicolinic Acid | Experimental (2) |
| 27 | Q0P9X4 | rpiB | Ribose 5-phosphate isomerase | Biosynthesis of secondary metabolites, Microbial metabolism in diverse environments | DB03661, DB03108, DB04496 | Cysteinesulfonic Acid, 4-Phospho-D-Erythronate, 4-Phospho-D-Erythronohydroxamic Acid | Experimental (3) |
| 28 | Q0PAZ1 | pbpB | Putative penicillin-binding protein | Peptidoglycan biosynthesis, beta-Lactam resistance | DB00274, DB00430, DB00438, DB01327, DB01328, DB01329, DB01331, DB01332, DB01414, DB01415, DB01598, DB04570, DB00319, DB00415, DB00456, DB00485, DB00493, DB00567, DB00607, DB00713, DB01603, DB08795, DB00303, DB08401, DB03313, DB08508, DB00739, DB01140, DB01163 | Cefmetazole, Cefpiramide, Ceftazidime, Cefazolin, Cefonicid, Cefoperazone, Cefoxitin, Ceftizoxime, Cefacetrile, Ceftibuten, Imipenem, Latamoxef, Piperacillin, Ampicillin, Cefalotin, Dicloxacillin, Cefotaxime, Cephalexin, Nafcillin, Oxacillin, Meticillin, Azidocillin, Ertapenem, (2e)-2-({(2s)-2-carboxy-2-[(phenoxyacetyl)amino]ethoxy}imino)pentanedioic acid, Cephalosporin C, N-benzoyl-D-alanine, Hetacillin, Cefadroxil, Amdinocillin | Approved (22); Approved, Investigational; Experimental (3); Approved, withdrawn (2); Withdrawn; |
| 29 | Q0PAL6 | pbpC | Penicillin-binding protein | Peptidoglycan biosynthesis, beta-Lactam resistance | DB01327 | Cefazolin | Approved |
| 30 | Q0PBA2 | frdC | Fumarate reductase cytochrome B subunit | Biosynthesis of secondary metabolites, Microbial metabolism in diverse environments, Two-component system | DB07669 | 2,3-dimethyl-1,4-naphthoquinone | Experimental |
| 31 | Q0P8D0 | ccoP | Cbb3-type cytochrome c oxidase subunit | Two-component system | DB02949, DB03224 | 2-Acetyl-Protoporphyrin Ix, 2-Formyl-Protoporphryn Ix | Experimental (2) |
| 32 | Q0PBM2 | cheA | Chemotaxis histidine kinase | Two-component system, Bacterial chemotaxis | DB02524, DB03909, DB04395 | Spiro(2,4,6-Trinitrobenzene[1,2a]-2o',3o'-Methylene-Adenine-Triphosphate, Adenosine-5′-[Beta, Gamma-Methylene]Triphosphate, Phosphoaminophosphonic Acid-Adenylate Ester | Experimental (3) |
| 33 | Q0PBM1 | cheV | Chemotaxis protein | Two-component system, Bacterial chemotaxis | DB02355, DB02596, DB07706, DB02461, DB03487, DB04156 | Adenosine-5′-Rp-Alpha-Thio-Triphosphate, Alpha,Beta-Methyleneadenosine-5′-Triphosphate, 2,3,17beta-trihydroxy-1,3,5(10)-estratriene, S-Methyl Phosphocysteine, 3-Aminosuccinimide, Aspartate Beryllium Trifluoride | Experimental (6) |
| 34 | P0C635 | cheY | Chemotaxis protein | Two-component system, Bacterial chemotaxis | DB02461, DB03487, DB04156, | S-Methyl Phosphocysteine, 3-Aminosuccinimide, Aspartate Beryllium Trifluoride, | Experimental (3) |
Discussion
The advent of a huge array of omics technologies has revolutionized the identification of drug targets (Russell et al., 2013). Examples include proteome scale comparative modeling of Corynebacterium pseudotuberculosis responsible for ulcerative lymphangitis, mastitis in ruminants (Hassan et al., 2014); RNA-seq profiling based target identification for non-small cell lung cancer (Riccardo et al., 2014); identification of markers for early prediction of preeclampsia using metabolic profiling (Kuc et al., 2014); computational systems biology approach for drug prioritization in Clostridium botulinum (Muhammad et al., 2014); and using protein–protein interaction network for drug target identification (Li et al., 2015).
The subtractive genomics approach is very efficient and has greatly accelerated the identification of relevant drug targets against many pathogens such as M. tuberculosis and Bacillus anthracis (Hosen et al., 2014; Rahman et al., 2014). Large-scale genomic projects such as the 1000 Genome and Encode are a rich source of background information and provide a deeper understanding of the genomes in relation to diseases. These computational approaches, together with the availability of genomic sequences, make it feasible for us to perform subtractive genomics and comparative pathway analysis aimed at identifying putative therapeutic drug targets and potential vaccine candidate proteins of C. jejuni.
Selecting drug targets using subtractive genomics approaches essentially relies on looking for those proteins that are absent in host but present in the pathogen, as it will minimize any adverse effects on host biology (Butt et al., 2012). Gene essentiality is also thought to be an important criteria for identification of therapeutic drug targets (Agüero et al., 2008). But there is a limitation to this approach as it fails to identify some targets such as hypoxanthine phosphoribosyl transferase as essential in Plasmodium falciparum (false negative) (Winzeler et al., 1999), while sometimes it yields false positives as in the case of dihydrofolate reductase in Leishmania major (Titus et al., 1995).
Gene essentiality prediction via experimental methods such as single gene knockouts, RNA interference, and conditional knockouts is labor-intensive, expensive, and time-consuming (Butt et al., 2012). Furthermore only a few infectious agents are amenable to experimental approaches of gene essentiality, as the tools for identification of drug or vaccine targets are often limited or absent for many pathogens. In such scenarios, computational methods for gene essentiality prediction seem to streamline the gap between the amount of data generated from sequencing projects and whole genome approaches for prediction of essential genes (Agüero et al., 2008; Doyle et al., 2010; Volker and Brown, 2002).
A previous work also identified the drug targets in C. jejuni by utilizing the CAI (Codon Adaptation Index) criterion as a measure of gene essentiality (Tilton et al., 2014) Essential genes are highly conserved, highly expressed, and preferentially positioned in the leading strand. But high gene expression rates do not correlate significantly with the gene strand biasness and nonessential genes also show high expression rates (Perrière and Thioulouse, 2002; Rocha and Danchin, 2003), in which case the definition of essentiality based on high expression could be erroneous. In our study, we have utilized a subtractive genomics approach to predict genes essential to C. jejuni through homology search against experimentally predicted essentiality data from C. jejuni and H. pylori in DEG, both of which belong to the epsilon class of proteobacteria.
Significant advancements in genome sequencing and bioinformatics coupled with experimental data have shown that factors which are determinant of structural and molecular properties of proteins, such as molecular weight, subcellular localization, transmembrane prediction, and availability of 3D structure, can aid in prioritization of drug targets (Agüero et al., 2008) and maximize the likelihood of landing to the best therapeutic target against pathogen, considerably reducing the time and resources for developing such an agent.
Molecular mass prioritization identified all the proteins with weight <110 kDa; suggesting the possibility of experimental verification of the identified targets. Smaller proteins are easy to purify, and localization information of proteins can yield insights into protein function (Duffield et al., 2010; Yu et al., 2010). There were only few experimentally solved structures available in PDB, which points to a gap in the structural characterization of pathogen proteins, albeit C. jejuni first whole genome was published in 2000 (Parkhill et al., 2000). Protein structural information can be used to a significant advantage in drug identification and validation, greatly reducing the cost of high throughput experimental assays (Grant, 2009). Sixteen proteins predicted as transmembrane represent potential vaccine candidate proteins, which were further filtered based on antigenicity and MHC-binding criteria.
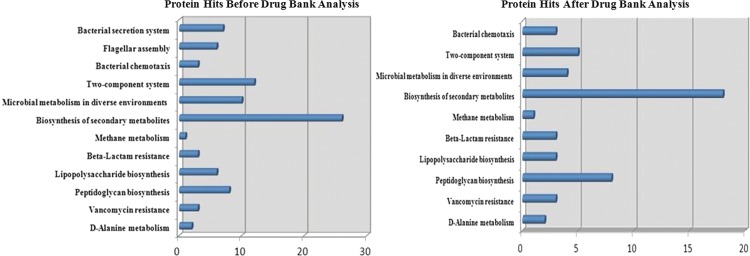
The likelihood of developing a drug-like compound to modulate the target is an important consideration that can aid drug design and an important determinant for a non-homologous protein to be a potential therapeutic target. The proteins for which drugs are already available can be useful starting points for drug discovery. The distribution of the essential non-homologous proteins was checked before and after similarity search against DrugBank (Fig. 2). It was noticed that there was ∼49% reduction in the number of proteins and 20% reduction in the number of pathways after DrugBank analysis, which leads to a shift in the pathway priority. Before DrugBank analysis biosynthesis of secondary metabolites, two-component system, microbial metabolism in diverse environments, peptidoglycan biosynthesis, bacterial secretion system, flagellar assembly, and lipopolysaccharide biosynthesis pathways were having maximum drug-able targets, but after DrugBank analysis, biosynthesis of secondary metabolites, peptidoglycan biosynthesis, two-component systems were the major pathways of drug-able targets.
FIG. 2.
Distribution of essential non-homologous proteins of C.jejuni in different pathways before and after drug bank search for drug-ability potential evaluation.
Finally, we have narrowed down the search of therapeutic targets to final 14 prioritized drug targets and vaccine candidates. 5′-Methylthioadenosine/S-adenosylhomocysteine nucleosidase (pfs) enzyme catalyzes the direct conversion of aminodeoxyfutalosine (AFL) into dehypoxanthine futalosine (DHFL) and adenine via the hydrolysis of the N-glycosidic bond that represents an essential step in the menaquinone biosynthesis pathway (Li et al., 2011). This enzyme is an attractive drug target as it is involved in many pathways, such as ubiquinone and other terpenoid-quinone biosynthesis, cysteine and methionine metabolism, and biosynthesis of amino acids.
Alanine racemase (alr) catalyzes the pyridoxal 5′-phosphate-dependent interconversion of L-alanine and D-alanine. D-alanyl-alanine synthetase A (ddl) is involved in cell wall formation by joining two of the D-alanine residues together, catalyzing the formation of the ATP-dependent D-alanine-D-alanine dipeptide bond between the resulting D-alanine molecules. Inhibition of these two enzymes leads to effective inhibition of peptidoglycan synthesis; ddl has been proposed to be an attractive drug target in Mycobacterium tuberculosis (Prosser and de Carvalho, 2013). Vancomycin binds to ddl, a peptidoglycan precursor, forming a stable complex under normal conditions and inhibits cell wall synthesis (Howden et al., 2010), ultimately leading to cell lysis. Thus ddl plays an important role in the vancomycin resistance pathway.
Penicillin Binding Proteins (PBPs) are of special interest, as these are target sites for beta-lactam antibiotics. They also play an important role in cell wall formation. pbpA is important for cell division and essential for growth (Wada and Watanabe, 1998). pbpB also is critical for bacterial growth and cell wall biosynthesis (Pinho et al., 2001). pbpC is a major protein of a cell division complex. PBPs have already been utilized as model drug target system (von Rechenberg et al., 2005). PBPs are highly similar to the binding partners of many FDA-approved and experimental drugs. Hence PBPs can be considered of high potential for experimental validation as vaccine candidates.
UDP-N-acetylglucosamine 1-carboxyvinyltransferase (murA) and UDP-N-acetylenolpyruvoylglucosamine reductase (murB) both catalyze important reactions in peptidoglycan precursor synthesis. murA catalyzes transfer of an enolpyruvate residue from phosphoenolpyruvate (PEP) to position 3 of UDP-N-acetylglucosamine, followed by a MurB-catalyzed NADPH dependent reduction of the UDP-N-acetylglucosamine enolpyruvate to UDP-N-acetylmuramic acid. Majority of antibiotics in clinical use today target later steps of peptidoglycan synthesis (El Zoeiby et al., 2003). murB is essential in Escherichia coli (Pucci et al., 1992). murA to murF genes are all essential and highly conserved among bacterial species, thus holding a great promise as future therapeutic drug targets.
Both frdC (fumarate reductase cytochrome B subunit) and ccoP are important constituents of oxidative phosphorylation pathway for ATP formation, often called a molecular unit of energy transfer. While frdC couples the reduction of fumarate to succinate with the oxidation of quinol to quinine, ccoP (Cbb3-type cytochrome c oxidase subunit) is required for transfer of electrons from donor cytochrome c via its heme groups to CcoO subunit. secD (protein translocase subunit SecD) a part of the Sec protein translocase complex, tatC (Sec-independent protein translocase protein TatC) an important part of the twin-arginine translocation (Tat) system and Cj1094c (Putative preprotein translocase protein) transports large proteins across membranes. The twin-arginine translocation (TAT) pathway is important to bacterial growth and virulence (Ding and Christie, 2003; Lavander et al., 2007). secD and Cj1094c help in secretion across the inner membrane via preprotein translocase pathway. Transport proteins are associated with pathogenesis and virulence and have been identified as potential vaccine candidates in several previous studies as well (Garmory and Titball, 2004; Harris et al., 2011).
Peptidoglycan is an important component of bacterial cell wall, responsible for maintaining a definite cell shape and primarily conferring mechanical resistance to higher osmotic pressure (Vollmer et al., 2008). Any interference with peptidoglycan biosynthesis will result in cell lysis. Peptidoglycan biosynthesis, the pathway with largest distribution of final identified drug targets, can be exploited for therapeutic drug targets owing to its multiple target enzymes whose inhibition could lead to disruption of cell well and in turn attenuate bacterial cell growth.
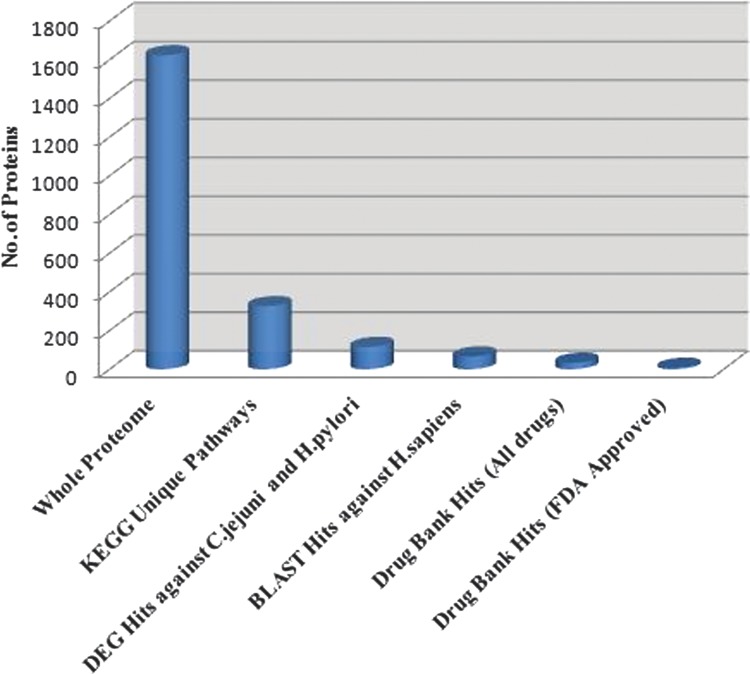
In summary, the computational subtractive genomics approach integrated with comparative pathway analysis resulted in a significant reduction in the number of protein targets (Fig. 3) at each step. Thus we were able to identify several essential proteins critical for bacterial growth and survival and with minimum toxicity to host. Further studies are warranted to validate these findings by in vitro and in vivo experiments for effective drug design against Campylobacter infections.
FIG. 3.
Step-wise reduction in the total number of proteins in the subtractive analysis of C.jejuni proteome for drug target identification.
Conclusion
We have performed subtractive genomics analyses of the C. jejuni pathogenic strain NCTC11168, and have identified several proteins in the genome that can prove to be potential targets for effective drug design. As many of the identified drug targets have already been reviewed to play critical role in the metabolic pathways that regulate bacterial growth and survival, a systematic approach to develop antibiotics against the identified targets would likely be very promising for the treatment of Campylobacter infections. Information about these targets can also lead to significant progress in testing the efficacy of already existing drugs, which is as equally important as development of new drugs. It is believed that the drugs developed against these identified targets will be pathogen specific and with minimal toxic effects on the host. Omics-guided drug discovery and bioinformatics analyses offer a broad and veritable potential for advances in global health relevant novel therapeutics (Cuadrat et al. 2014; Preidis and Hotez, 2015).
Acknowledgments
This research was supported by Fast Track Young Scientist Fellowship from DST (Department of Science and Technology), Ministry of Science and Technology, India under grant number SB/FT/LS-278/2012.
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Agüero F, Al-Lazikani B, Aslett M, et al. (2008). Genomic-scale prioritization of drug targets: The TDR Targets database. Nat Rev Drug Discov 7, 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar SQ. (1988). Antimicrobial sensitivity and plasmid-mediated tetracycline resistance in Campylobacter jejuni isolated in Bangladesh. Chemotherapy 34, 326–331 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, et al. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, et al. (2000). The Protein Data Bank. Nucleic Acids Res 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AM, Nasrullah I, Tahir S, et al. (2012). Comparative genomics analysis of Mycobacterium ulcerans for the identification of putative essential genes and therapeutic candidates. PLoS One 7, e43080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. (2000). Preliminary FoodNet data on the incidence of foodborne illnesses—Selected sites, United States, 1999. MMWR. Morb Mortal Wkly Rep 49, 201–205 [PubMed] [Google Scholar]
- CDC. (2013). Antibiotic Resistance Threats in the United States, 2013
- Chawley P, Samal HB, Prava J, et al. (2014). Comparative genomics study for identification of drug and vaccine targets in Vibrio cholerae: MurA ligase as a case study. Genomics 103, 83–93 [DOI] [PubMed] [Google Scholar]
- Cuadrat RR, da Serra Cruz SM, Tschoeke DA, et al. (2014) An orthology-based analysis of pathogenic protozoa impacting global health: An improved comparative genomics approach with prokaryotes and model eukaryote orthologs. OMICS 18, 524–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, and Christie PJ. (2003). Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J Bacteriol 185, 760–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MA, Gasser RB, Woodcroft BJ, et al. (2010). Drug target prediction and prioritization: Using orthology to predict essentiality in parasite genomes. BMC Genomics 11, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova IA, and Flower DR. (2007). VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformat 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield M, Cooper I, McAlister E, et al. (2010). Predicting conserved essential genes in bacteria: In silico identification of putative drug targets. Mol Biosyst 6, 2482–2489 [DOI] [PubMed] [Google Scholar]
- El Zoeiby A, Sanschagrin F, and Levesque RC. (2003). Structure and function of the Mur enzymes: Development of novel inhibitors. Mol Microbiol 47, 1–12 [DOI] [PubMed] [Google Scholar]
- Engberg J, Aarestrup FM, Taylor DE, et al. (2001). Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: Resistance mechanisms and trends in human isolates. Emerg Infect Dis 7, 24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, et al. (1995). Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512 [DOI] [PubMed] [Google Scholar]
- Fraser CM, Gocayne JD, White O, et al. (1995). The minimal gene complement of Mycoplasma genitalium. Science 270, 397–403 [DOI] [PubMed] [Google Scholar]
- Garmory HS, and Titball RW. (2004). ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 72, 6757–6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Prava J, Samal HB, et al. (2014). Comparative genomics study for the identification of drug and vaccine targets in Staphylococcus aureus: MurA ligase enzyme as a proposed candidate. J Microbiol Methods 101, 1–8 [DOI] [PubMed] [Google Scholar]
- Grant MA. (2009). Protein structure prediction in structure-based ligand design and virtual screening. Comb Chem High Throughput Screen 12, 940–960 [DOI] [PubMed] [Google Scholar]
- Haas J, Roth S, Arnold K, et al. (2013). The Protein Model Portal—A comprehensive resource for protein structure and model information. Database (Oxford) 2013, bat031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JA, Roy K, Woo-Rasberry V, et al. (2011). Directed evaluation of enterotoxigenic Escherichia coli autotransporter proteins as putative vaccine candidates. PLoS Negl Trop Dis 5, e1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan SS, Tiwari S, Guimarães LC, et al. (2014). Proteome scale comparative modeling for conserved drug and vaccine targets identification in Corynebacterium pseudotuberculosis. BMC Genomics 15, S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoge CW, Gambel JM, Srijan A, et al. (1998). Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin Infect Dis 26, 341–345 [DOI] [PubMed] [Google Scholar]
- Hosen MI, Tanmoy AM, Mahbuba DA, et al. (2014). Application of a subtractive genomics approach for in silico identification and characterization of novel drug targets in Mycobacterium tuberculosis F11. Interdiscip Sci 6, 48–56 [DOI] [PubMed] [Google Scholar]
- Howden BP, Davies JK, Johnson PDR, et al. (2010). Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 23, 99–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, and Goto S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, et al. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol 305, 567–580 [DOI] [PubMed] [Google Scholar]
- Kuc S, Koster MPH, Pennings JLA, et al. (2014). Metabolomics profiling for identification of novel potential markers in early prediction of preeclampsia. PLoS One 9, e98540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavander M, Ericsson SK, Bröms JE, et al. (2007). Twin arginine translocation in Yersinia. Adv Exp Med Biol 603, 258–267 [DOI] [PubMed] [Google Scholar]
- Li X, Apel D, Gaynor EC, et al. (2011). 5′-methylthioadenosine nucleosidase is implicated in playing a key role in a modified futalosine pathway for menaquinone biosynthesis in Campylobacter jejuni. J Biol Chem 286, 19392–19398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZC, Zhong WQ, Liu ZQ, et al. (2015). Large-scale identification of potential drug targets based on the topological features of human protein–protein interaction network. Anal Chim Acta 871, 18–27 [DOI] [PubMed] [Google Scholar]
- Muhammad SA, Ahmed S, Ali A, et al. (2014). Prioritizing drug targets in Clostridium botulinum with a computational systems biology approach. Genomics 104, 24–35 [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668 [DOI] [PubMed] [Google Scholar]
- Perrière G, and Thioulouse J. (2002). Use and misuse of correspondence analysis in codon usage studies. Nucleic Acids Res 30, 4548–4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Davis FP, et al. (2006). MODBASE: A database of annotated comparative protein structure models and associated resources. Nucleic Acids Res 34, D291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho MG, de Lencastre H, and Tomasz A. (2001). An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci 98, 10886–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preidis GA, and Hotez PJ. (2015) The newest “omics”—metagenomics and metabolomics—enter the battle against the neglected tropical diseases. PLoS Negl Trop Dis 9, e0003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser GA, and de Carvalho LPS. (2013). Kinetic mechanism and inhibition of Mycobacterium tuberculosis D-alanine:D-alanine ligase by the antibiotic D-cycloserine. FEBS J 280, 1150–1166 [DOI] [PubMed] [Google Scholar]
- Pucci MJ, Discotto LF, and Dougherty TJ. (1992). Cloning and identification of the Escherichia coli murB DNA sequence, which encodes UDP-N-acetylenolpyruvoylglucosamine reductase. J. Bacteriol 174, 1690–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Noore S, Hasan A, et al. (2014). Identification of potential drug targets by subtractive genome analysis of Bacillus anthracis A0248: An in silico approach. Comput Biol Chem 52, 66–72 [DOI] [PubMed] [Google Scholar]
- Reina J, Ros MJ, and Serra A. (1994). Susceptibilities to 10 antimicrobial agents of 1,220 Campylobacter strains isolated from 1987 to 1993 from feces of pediatric patients. Antimicrob Agents Chemother 38, 2917–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardo F, Arigoni M, Buson G, et al. (2014). Characterization of a genetic mouse model of lung cancer: A promise to identify Non-Small Cell Lung Cancer therapeutic targets and biomarkers. BMC Genomics 15, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC, and Danchin A. (2003). Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res 31, 6570–6577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C, Rahman A, and Mohammed AR. (2013). Application of genomics, proteomics and metabolomics in drug discovery, development and clinic. Ther Deliv 4, 395–413 [DOI] [PubMed] [Google Scholar]
- Samal HB, Prava J, Suar M, et al. (2015). Comparative genomics study of Salmonella Typhimurium LT2 for the identification of putative therapeutic candidates. J Theor Biol 369C, 67–79 [DOI] [PubMed] [Google Scholar]
- Sarangi AN, Aggarwal R, Rahman Q, et al. (2009). Subtractive genomics approach for in silico identification and characterization of novel drug targets in Neisseria meningitides serogroup B J Comput Sci Syst Biol 2, 255–258 [Google Scholar]
- Singh H, and Raghava GPS. (2003). ProPred1: Prediction of promiscuous MHC Class-I binding sites. Bioinformatics 19, 1009–1014 [DOI] [PubMed] [Google Scholar]
- Tilton F, Priya L, Nair K, et al. (2014). Insilico metabolic pathway analysis for potential drug target in campylobacter jejuni | World Research Publications. Int J Healthc Pharm Res 3, 029–032 [Google Scholar]
- Titus RG, Gueiros-Filho FJ, de Freitas LA, et al. (1995). Development of a safe live Leishmania vaccine line by gene replacement. Proc Natl Acad Sci 92, 10267–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker C, and Brown JR. (2002). Bioinformatics and the discovery of novel anti-microbial targets. Curr Drug Targets Infect Disord 2, 279–290 [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, and de Pedro MA. (2008). Peptidoglycan structure and architecture. FEMS Microbiol Rev 32, 149–167 [DOI] [PubMed] [Google Scholar]
- Von RM, Blake BK, Ho YSJ, et al. (2005). Ampicillin/penicillin-binding protein interactions as a model drug-target system to optimize affinity pull-down and mass spectrometric strategies for target and pathway identification. Proteomics 5, 1764–1773 [DOI] [PubMed] [Google Scholar]
- Wada A, and Watanabe H. (1998). Penicillin-binding protein 1 of Staphylococcus aureus is essential for growth. J Bacteriol 180, 2759–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization 2011, WHO/Campylobacter, accessed May27, 2015 <http://www.who.int/mediacentre/factsheets/fs255/en/>
- Winzeler EA, Shoemaker DD, Astromoff A, et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, et al. (2008). DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 36, D901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, et al. (2010). PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Ou HY, and Zhang CT. (2004). DEG: A database of essential genes. Nucleic Acids Res 32, D271–272 [DOI] [PMC free article] [PubMed] [Google Scholar]