Abstract
An anti-malarial vaccine against the extremely lethal Plasmodium falciparum is desperately needed. Peptides from this parasite's proteins involved in invasion and having high red blood cell-binding ability were identified; these conserved peptides were not immun genic or protection-inducing when used for immunizing Aotus monkeys. Modifying some critical binding residues in these high-activi binding peptides' (HABPs') attachment to red blood cells (RBC) allowed them to induce immunogenicity and protection against expermental challenge and acquire the ability to bind to specific HLA-DRp1* alleles. These modified HABPs adopted certain characterist structural configurations as determined by circular dichroism (CD) and 1H nuclear magnetic resonance (NMR) associated with certain HLA-DRβ1* haplotype binding activities and characteristics, such as a 2-Å-distance difference between amino acids fitting into HLA-DRp1 Pockets 1 to 9, residues participating in binding to HLA-DR pockets and residues making contact with the TCR, suggesting haplotyp and allele-conscious TCR. This has been demonstrated in HLA-DR-like genotyped monkeys and provides the basis for designing high effective, subunit-based, multi-antigen, multi-stage, synthetic vaccines, for immediate human use, malaria being one of them.
Keywords: anti-malarial vaccine, P. falciparum, HLA-DRβ1* molecules
Introduction
P. falciparum invasion of RBCs
Merozoite proteins involved in invading erythrocytes
Erythrocyte proteins involved in merozoite invasion
The state of current worldwide anti-malarial vaccine approaches
A rational approach towards developing subunit-based synthetic vaccines
The immune response elicited by conserved HABPs
Structural analysis of native and modified HABPs
Secondary structure analysis
Native and modified HABP 3D structure explains some immunological phenomena
Supporting the haplotype – and allele-conscious TCR concept
Modified HABPs' 3D structure revealed a fit into HLA molecules
Conclusion
Introduction
Plasmodium falciparum is the most lethal malarial parasite, which attacks more than 500 million people and kills 2 million of them each year, mainly children aged less than 5 in Africa [1]. Control measures against this threatening disease are therefore desperately needed, such as development of vaccine and new anti-malarial drugs.
It has to be kept in mind when developing a totally effective vaccine against P. falciparum malaria that as suggested by transcrip-tome analysis, around 58–90 merozoite proteins are involved in this parasite's invasion of red blood cell (RBC) [2] and a similar number of sporozoite proteins could be acting in hepatocyte invasion [3].
Most merozoite-recognized proteins have been functionally and molecularly studied by others [4] as well as by ourselves [5].
Work by other groups has shown that their synthesis and expression can be switched on and off [6] (depending on parasite invasion requirements and/or immunological pressure [7]) and trimmed [8] or processed [9] to perform their biological functions.
A totally effective anti-malarial vaccine must therefore contain at least a similar number of merozoite and sporozoite proteins or epitopes representing these molecules' most relevant functional parts (multi-epitope, multi-stage) to ensure complete blocking of the parasite during its critical invasion stages [5, 10].
The first multi-epitope, multi-stage, subunit-based, anti-malarial vaccine, named SPf66, was developed by us 20 years ago [11, 12] following a large set of experiments in Aotus monkeys involving synthetic peptides. It showed that chemically synthesized vaccines were feasible, safe and immunogenic, providing complete protection for ∼40% of immunized monkeys and humans exposed to experimental challenge [11, 12]. It was ∼38.8% in humans in Colombia [13], 55.1% in Venezuela [14], 60.8% in Ecuador [15], 31% in Tanzania [16] for up to 2 years [17] in large field trials carried out on individuals aged more than 1 year [13, 16] in different ethnic and epidemiological set-ups around the world. SPf66 was ineffective in infants aged less than 1 [18] and a batch with a different degree of polymerization produced elsewhere was not protective [19], thereby dropping its protective efficacy in a recent meta-analysis [20].
SPf66 contained four epitopes; three of them were merozoite-derived, chemically synthesized, high-activity binding peptides (HABPs) attaching to RBC [21], and one a circumsporozoite protein-derived, having high hepatocyte-binding ability [12]. SPf66 taught us that a protection-inducing immune response should thus be directed against the parasite's functionally relevant amino acid sequences (i.e. HABPs), specifically conserved ones (to rule out the P. falciparum parasite's tremendous genetic polymorphism).
SPf66 also taught us the importance of the host's genetic variability linked to major histocompatibility (MHC)–peptide–T-cell receptor (TCR) complex formation, since non-responders and non-protected individuals typing HLA-DRβ1*04 [22] preferentially used TCR Vβ3 and Vβ11 families [23]. This experimental, clinical and field trial data clearly suggested that genetic control of HLA-DRβ1* allele-associated immune response had to be very seriously taken into account when developing vaccines. Here we briefly show that this approach is completely feasible.
Besides the parasite's genetic polymorphism, the human immune system's genetic polymorphism associated with controlling the immune response and dominated by major histocompatibility complex class II molecules (MHC II), particularity HLA-DRβ1* molecules, adds tremendous complexity to vaccine development. HLA-DRβ1* has two chains: the almost monomorphic alpha (a) chain encoded by the HLA-DRA region and the beta (β) chain encoded by the HLA-DRB region having wide genetic polymorphism [24].
Sixteen MHC class II allelic molecules (MHC II) encoded by the HLA-DR region have been defined (HLA-DRβ1*01–16); there are more than 250 variants [24]. These alleles can be serologically, functionally, evolutionally and molecularly grouped into five large groups or haplotypes (alleles are given in brackets) named HLA-DR1 (HLA-DRβ1*01, 10, 103, 104), HLA-DR51 (HLA-DRp1*15 and 16) HLA-DR52 (HLA-DRp1*03, 11, 12, 13 and 14), HLA-DR8 (only HLA-DRp1*08) and HLA-DR53 (HLA-DRp1*04, 07, 09) [25]. These molecules are able to present peptide antigens to T-cell receptors (TCR) when the antigen fits properly into its binding site, thereby forming the appropriate MHC II–peptide–TCR complex and inducing an optimal immune response.
Clinical studies related to anti-malarial vaccine development (especially Phase III trials) are surrounded by a high degree of uncertainty owing to the large number of confounding factors such as the individual's and parasite's genetic variability; lack of knowledge of the immunological principles determining a protection-inducing immune response; the amount of inoculum injected during P. falciparum-infected Anopheles mosquito bites; the physical, chemical and biological characteristics of the antigens used for inducing immune protection and the variability of the vaccine batches being used for immunoprophylaxis.
The Aotus monkey represents an appropriate experimental model for solving these problems in human anti-malarial vaccine development owing to its extreme susceptibility to human malaria [26, 27] and the reproducible results obtained when intravenously infected with the parasite's erythrocyte stages (infected RBCs), meaning that the amount of inoculum can be precisely established and the disease's evolution can be clearly quantified [26, 27]. This monkey's immune system molecules display very high similarity with those of humans at the nucleotide and amino acid sequence level, such as immunoglobulins, cytokines and CD molecules, making this primate an excellent model for developing anti-malarial vaccines for use in humans.
Sequencing studies of the genes encoding these Aotus Class II molecules (HLA-DRβ1*-like), undertaken in 110 Aotus monkeys, have revealed 100–88% homology with human HLA-DRβ1*04, 03, 08, 11, 13, 14, 15, 16, 10, 07 and 01 molecules [28] and >80–100% homology with human TCRs [29] and other immune-system molecules [30].
It has been reported that these HLA-DRβ1*-associated genes' allele frequency in Aotus is ∼25% for HLA-DRβ1*0403/0407, HLA-DRβ1*0422, HLA-DRβ1*0301, HLA-DRβ1*15–16 alleles, 10% for HLA-DRβ1*08–11, 7% for HLA-DRβ1*1001 and HLA-DRβ1* 0101 and 4% for HLA-DRβ1*0701 [28]. It is not surprising then that the maximum protective efficacy which a synthetic vaccine subunit can induce is that of the frequency of the allele to which it binds (∼25%).
Molecular biology analysis of Aotus MHC-DRB1-like proteins and all other immune system molecules could thereby lead to the logical and rational design of vaccines that could be recognized by both Aotus and human HLA-DRB_molecules. These could then be used immediately in humans, thus avoiding risky, costly, and prolonged clinical trials involving thousands of human beings simply to test a single molecule or a group of them, accompanied by all their inherent ethical, scientific and logistical problems.
A group of leading scientists attending a meeting sponsored by the Academy of Medical Sciences, the Royal Society, the Medical Research Council and the Wellcome Trust recently concluded that, ‘even with the money pouring in from people like Bill Gates, the vast cost of large-scale clinical trials meant that the world could not afford to squander money for research into AIDS and human vaccines like AIDS, malaria and tuberculosis’, since ‘pre-testing in a small number of non-human primates can ensure we proceed into human trials with vaccines that are likely to succeed’, [31] an approach fiercely promoted by our Institute for almost the last three decades.
A thorough 1H-NMR study of the three-dimensional (3D) structure of epitopes that could be included in a vaccine was thus undertaken in our institute 14 years ago, specifically aimed at synthetic vaccine development. The structural analysis of HABPs or their analogues inducing a protective immune response against malaria was thus started to gain a deep understanding of the physicochemical rules making a vaccine protective and effective. This was in an attempt to find a structure–function relationship and to define mathematical, physical, chemical and biological rules for vaccine development, malaria being the target for one such vaccine and this study's raison d être.
P. falciparum invasion of RBCs
P. falciparumis one of four malarial species that are infectious for humans. It is transmitted by an infected female Anophelesmos-quito bite injecting larvae-like structures (sporozoites) with her saliva during a blood meal. The sporozoites are transported with the blood and migrate into hepatocytes in the liver, where they replicate 30,000 times in a week and differentiate into pear-like structures (merozoites), which are released into the host's bloodstream [32].
The merozoite rolls over the RBC surface seeking new erythro-cytes to invade during the asexual or erythrocyte cycle. Receptor-ligand interactions between merozoite proteins and the different molecules located on the RBCs lead to merozoite binding and reorientation on RBC in less than 40 sec., ending by being detained on some of these RBCs to begin binding [33]. The merozoite brings its apical pole into contact and anchors to the RBC membrane via the apical merozoite antigen-1 (AMA-1) molecule, rearranging its microneme proteins on its surface and releasing others from the rhoptries onto the RBC membrane, acting as bridges between the two cells. Merozoite reorientation is associated with erythrocyte deformation to facilitate merozoite penetration and formation of the parasitophorous vacuole (PV) [34].
Some other rhoptry and dense granule proteins, such as ring erythrocyte surface antigen (RESA) family proteins, are released when this large, round PV is formed, after which the cytoplasm and a ring-like structure or ring stage appears in the chromatin granule forming it.
It develops into a mature form (trophozoite) as the process continues where the parasitic machinery now becomes fully involved in the parasite's molecular synthesis. Its nucleus then divides to form early schizonts, giving rise (a few hours later) to mature or late schizonts that burst, thereby releasing new merozoites that invade the new RBC as part of a cycle in which host cells are killed off [32–34].
Some merozoites can also change their morphology to produce male and female gametocytes in a cycle called gametogenesis. A mosquito biting a human being infected with the parasite receives gametocyte-infected erythrocytes [35], thereby starting the reproductive sexual cycle in the mosquito's midgut.
Merozoite proteins involved in invading erythrocytes
Transcriptome analysis has shown that 58–90 merozoite proteins are implicated in the multi-step invasion of erythrocytes [2] as they are so critical for parasite survival. Some of these proteins are situated on the merozoite surface, whilst others are located in the rhoptries, micronemes and dense granules (Fig. 1). These proteins have been grouped into several families according to their cellular location or molecular characteristics.
Fig 1.

The merozoite structure, with its most important cellular components (apical pole, rhoptries, micronemes, dense granules) and molecules localized in these organelles involved in merozoite invasion of RBC.
The merozoite surface protein family (MSP1 to MSP10) represents a group of proteins situated on the merozoite surface (Fig. 1). MSP 1, 2, 4, 5, 8 and 10 are anchored to the membrane via glycosylphosphatidylinositol (GPI) tails [36–39] (Fig. 1) and it has been suggested that they are involved in the initial steps of merozoite invasion of RBC.
Other MSP proteins have transmembrane domains, whilst still others are soluble, such as serine repeat antigen (SERA) [40], or may be weakly bound to the merozoite surface or associated with other membrane proteins like MSP-3, -6 and -7 [41–43] which are bound to MSP-1 [44] forming large macromolecular complexes [9] or lipid rafts during invasion (Fig. 2). Some MSP family molecules contain one or two epidermal growth factor (EGF)-like domains [45], suggesting that protein–protein interactions may be involved in merozoite invasion.
Fig 2.

Macromolecular complexes of merozoite proteins involved in RBC invasion present on detergent-resistant membrane (DRM), DRM-associated lipid rafts as recognized in DRM proteomes. Molecule sizes are drawn at their approximate molecular weights. Some molecules remain anchored to the parasites' DRM-associated lipid rafts via their GPI tails (shown as black twists traversing the pale blue membrane) such as MSP-1 (golden), MSP-2 (light blue), MSP-4 (pink), MSP-5 (dark blue), Pf 92 (brown), Pf 12 (clear purple) and RAMA (purple). Some other molecules are non-covalently bound, like RAP-1, -2 and -3 molecules to RAMA and MSP-6 (fuchsia) and -7 (green) to MSP-1. All members of the high (Rhop H) molecular weight rhoptry proteins having transmembrane sequences were also identified in proteome analysis. Another raft is formed by MSP-1 82-kD, 30-kD, 38-kD fragments (gold) and MSP-1 33-kD (red) and the 19-kD (red) fragment anchored to the merozoite membrane via the GPI tail to which some other molecules bind non-covalently, as shown in the central portion of this figure. (A) front and (B) top views.
Another merozoite protein family involved in invasion (located in the micronemes) (Fig. 1) displays similar internal structural composition, having a cysteine-rich inter-domain region (CIDR) located in these molecules' N-terminal portion generally involved in erythrocyte-binding activity, an intermediate region, another cysteine-rich region located in the C-terminal portion, a trans-membrane region and a small cytoplasmatic tail. This protein family displays a good number of frequently conserved cysteines and aromatic residues. They have been called erythrocyte-binding ligands(EBLs) as they are able to bind to different receptors on the erythrocyte membrane [46]. This protein family has been grouped into the so-called Duffy binding-like family (DBL family), P. falciparumerythrocyte-binding antigen-175 (EBA-175) [47], EBA-140 [48], EBA-181 [49], EBA-165 and EBL-1 [50], forming part of this family due to structural similarities with P. vivax and P. knowlesi protein binding to the Duffy receptor.
Another protein family located in the rhoptries that could be playing an important role in merozoite apical interaction and subsequent biochemical events is called the reticulocyte-binding-like (RBL) family, including normocyte-binding proteins (NBP-1, NBP-4), reticulocyte-binding proteins homologous-1 (RBP-H1), RBP-2-Ha, RBP-2-Hb, rhoptry-associated protein (RAP) 1 to 3, rhoptry proteins (RHOP) H1, H2, H3 and 148, the cytoadherence-like asexual gene (CLAG) family 1 to 9 [51, 52] (Fig. 1).
Most of these proteins organize themselves into detergent-resistant membrane (DRM)-associated macromolecular complexes where some of them, anchored to the membrane viaGPI tails (MSP-1, -2, -4, -5 and RAMA) [53, 54], serve as support for others that are loosely bound (MSP-6, -7, Pf41) for mediating coordinated interaction with RBCs (Fig. 2).
These molecules allow the junction to be formed and merozoite interaction with erythrocytes before the cellular complex becomes irreversibly bound. Many more proteins have been involved in merozoite invasion of RBC [44] but only the most relevant in this are mentioned.
Erythrocyte proteins involved in merozoite invasion
Merozoite invasion of erythrocytes is mediated by molecular communication between both types of cell. Merozoites must “persuade” erythrocytes to help in forming the PV membrane so that successful invasion can occur. Taking the erythrocyte cytoskeleton apart or destabilizing it occurs so that the intracellular vacuole can be formed; it thus becomes one of the main biochemical events occurring during invasion.
The merozoite must send the information from outside the erythrocyte, penetrating the erythrocyte membrane via the trans-membrane proteins that interact with the RBC cytoskeleton, such as sialoglycoproteins (i.e. glycophorin A, B and C) [55] and band 3. It has been reported that glycophorins A and B interact with merozoite protein erythrocyte-binding antigen 175 (EBA-175), glycophorin C with EBA-140 protein and band 3 with a merozoite surface protein 1 fragment (MSP-1) called MSP-142 and acid basic repeat antigen (ABRA) protein (MSP-9). Evidence has also been presented that glycophorins mediate initially weak, relatively nonspecific merozoite binding to RBC, where net glycophorin load is very important [56–58].
Enzyme treatment of different RBCs has been described, as have binding assays involving different antigens and receptor X. P. falciparum normocyte binding protein 1 (PfNBP1) binds to erythrocytes, such adhesion being dependent on a neuraminidase-sensitive, trypsin-resistant Z receptor. P. falciparum reticulocyte homologue 2b protein (PfRH2b) ligand mediates invasion through a chymotrypsin-sensitive, trypsin/neuraminidase-resistant receptor, which has also been referred to as receptor Z [59].
The state of current worldwide anti-malarial vaccine approaches
More than 40 assays in humans summarized by Engers and Godal by 1998 [60] using other molecule candidates for an anti-malarial vaccine have led to frustrating results. The situation has not improved too much during the last 10 years.
A vaccine's efficacy in conferring close to 100% individual protection is needed for studying the parasite's pre-erythrocyte (sporozoite) stage as, if just one sporozoite escapes, the immune response to this can lead to the complete development of the disease.
Studies of pre-erythrocyte stage synthetic vaccines consisting of unmodified circumsporozoite protein (CSP) amino acid sequences, based on its tandem repeat sequences or elongated with T helper epitopes [61], have given negative results.
Totally negative and frustrating results have also been reported for DNA fragments encoding thrombospondin-related protein (TRAP) sporozoite amino acid sequences + CSP and inserted into genetically modified vectors such as fowl pox (FP9) and the modified vaccinia virus (MVA) and long DNA fragments corresponding to multiple genes encoding an increasing number of proteins from the parasite's pre-erythrocyte and blood stages [62–64].
The RTS,S/AS02A is the only sporozoite-stage vaccine candidate that has provided ∼30% protection for up to 11/2 years. This is a P. falciparum circumsporozoite protein (CSP) recombinant fusion protein (amino acids 283–384) integrated with a hepatitis B virus surface antigen, to which a group of potent immunopoten-tiators has been added, such as muramil-dipeptide (MDP), the saponin-derived QS-21, ASO [65].
Equally negative results have been reported for anti-blood stage (merozoite) vaccines, which have involved different strategies ranging from immunization with ultra-low doses of the parasite [66] to virosome-based, multi-antigen peptide vaccines [67]. Merozoite-stage vaccine NYVAC-7 (containing genes encoding seven molecules derived from different parasite proteins) protected 1 of 35 vaccinated volunteers against experimental challenge and was thus discarded owing to its low protection-inducing ability (<3%) [68]; the same happened to MUST DO 7.
A mixture of MSP1, MSP2 and RESA recombinant proteins that was tested in field trials in Papua-New Guinea did not produce a statistically significant degree of protection in the vaccinated population [69]. This led to frustrating results, as challenging non-immune participants did not reveal any reduction or delay in parasite growth rates.
Another vaccination assay involving individual recombinant proteins or their fragments (such as the 42-kD MSP1 called FMP1) led to equally frustrating results when tested on a small group of children in Kenya (phase IIb field-trial) (unpublished data, 2006). Combining both MSP-2 recombinant allele forms tested in Papua-New Guinea returned similarly negative field trial results. Assays with AMA-1 protein have been equally frustrating. None of the other recombinant proteins, such as MSP-3 and SERA, or mixtures of GLURP + MSP 3, 19-kD MSP-1 + AMA 1, AMA-1 virosomes + CSP or EBA-175 have gone to more than phase I (safety and immunogenicity) clinical trials.
When any of these molecules has been used as a vaccine it may have been highly immunogenic, a cytotoxic T-lymphocyte inducer and have even been taken from antibodies in a few individuals, but their protection-inducing ability has been null or very low.
SPf66 has therefore been the first successful vaccine and the first and only one against malaria tested in large human field trials showing a high degree of protective efficacy against this deadly disease.
It should also be stressed that all immune responses induced by other vaccine candidates that have been found have been species- and strain-specific, reinforcing our findings that all immune responses must be directed against conserved HABPs.
A rational approach towards developing subunit-based synthetic vaccines
A methodology for identifying these molecules' most relevant parts, regions or amino acid sequences involved in invasion was first developed at our institute 15 years ago. It was aimed at recog-nizing merozoite- and sporozoite-HABPs involved in invading RBCs and hepatocytes and their critical binding residues recognized via glycine analogue scanning [70, 71].
Approximately 20-mer-long peptides, covering the whole length of known merozoite proteins involved in RBC invasion, have thus been synthesized in our institute and tested in erythrocyte-binding assays. Experimental binding assay conditions (defined from theoretical binding curves using a bimolecular interaction) were chosen for identifying HABPs recognizing 1,000 to 100,000 binding sites per cell. Specifically bound peptides' relationship to added peptide is directly proportional to the affinity constant and number of peptide receptors on erythrocytes or host cells (binding ability) in these conditions [70, 71].
This can be defined by:
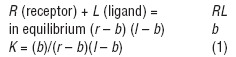 |
Where r = peptide receptor concentration, l = peptide concentration, b = bound peptide concentration and K= affinity constant. The (b)/(l) ratio is defined as the binding activity; those peptides having binding activity greater than or equal to 0.02 (2%) are considered to have high binding activity and are named high-activity binding peptides (HABPs), indicating that there are more than 2,000 receptor sites per cell [70–72]. Figure 3 shows the amino acid sequences of EBA-175 [71], MSP-2 [73], AMA-1 [74] and histidine-rich protein II (HRP-II) [75]. The black bars show their corresponding binding activity.
Fig 3.
An example of amino acid sequence and specific human RBC binding activity for chemically synthesized EBA-175 [71], MSP-2 [73], AMA-1[74] and HRPII protein [75] peptides is displayed. The peptides, with their corresponding amino acid sequences, are given on the left-hand side; small numbers indicate their position within the native protein. The peptide number given is the code assigned by our laboratory for each peptide used. The dotted line to the right separates peptides having >2% binding ability. Each peptide's specific binding activity is indicated by the black bar; 2% binding ability in the screening binding assay represents ∼1200 binding sites per cell. The dark grey area corresponds to amino acid sequences having little or no genetic variability. HABPs localized in these regions are called conserved HABPs; the others, located in the clear areas, are named variable HABPs.
Binding assay conditions have been standardized, mainly bearing factors in mind that could affect merozoite invasion of erythrocytes, such as haematocrit level, reaction time and temperature, leading to a highly sensitive, specific and robust methodology for identifying these HABPs [70, 71].
Most of our studies have shown that most HABPs bind to human erythrocyte membrane molecules, since ≥60% of HABP erythrocyte-binding ability can be removed by enzymatic treatment with neuraminidase, trypsin or chymotrypsin [76].
HABP-binding constants have also been determined via saturation assays in which ligand concentration is kept constant in the presence of growing concentrations of radioactively labelled ligand. Affinity constants (Kd), maximum number of binding sites (Bmax) and Hill coefficients (nH) have been obtained in these conditions for all HABPs found to date. HABP binding to erythrocytes is saturable (Fig. 2A), has simple interaction characteristics, 60–1100 nM affinity constants and 2,000–100,000 binding sites per cell. Hill plots have revealed that most of these peptides display positive cooperativity, suggesting that once a ligand has bound to its receptor site, it facilitates the next ligand's binding [70–80].
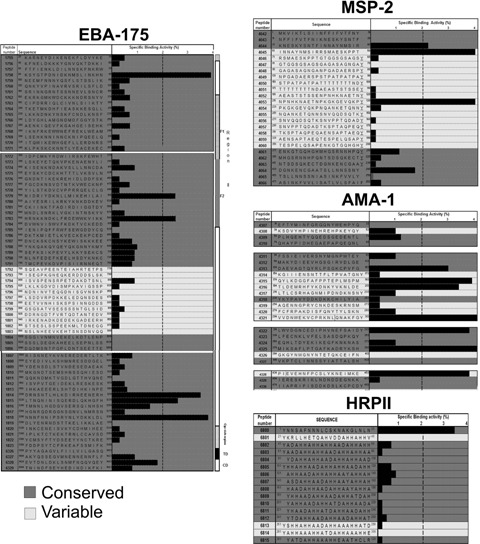
Critical residues in HABP binding to receptor cells have been determined for a large number of these HABPs by competition assays between HABP and glycine analogues as shown for AMA-1, HABPs 4313, 4325 and 4337 [74] (Fig. 4). Critical residues are those where peptide analogues show at least 50% reduced specific binding in their ability to compete with original peptide at three logarithmic concentrations: 10, 100 and 800 nM.
Fig 4.
An example of competition binding assays with 4313, 4325 and 4337 conserved HABP analogues (AMA-1 derived) [74]. Original radioisotope-labelled peptide-specific binding inhibited by analogous peptide is shown at 100 and 800 nM. The inhibition assay was performed with original non-labelled peptides and their analogous peptides. The asterisk (*) represents original peptides' specific binding. Underlined residues corresponding to the critical binding residues, whose modifications were analysed at 10, 100 and 800 nM concentrations (only two are shown in the figure), displayed the same reduced RBC binding pattern (≥50%) when modified.
The immune response elicited by conserved HABPs
Some HABP sequences are targets for antibodies inhibiting the invasion of erythrocytes. Immunogens containing part of an HABP, or peptide sequences analogous to that of an HABP, induce antibodies that have inhibited merozoite invasion and also induced protection in an experimental animal model [81]. However, these HABPs have displayed high variability in their amino acid sequences (variable HABPs) within different P. falciparum strains or isolates, making their use highly improbable owing to the huge number of variants against which the immune response should be directed, with all the inherent methodological and immunological problems. Attention has thus been directed against those having no genetic variation.
However, HABPs that do not present genetic variability in their amino acid sequences in different isolates or strains from different parts of the world (conserved HABPs, shown in the grey areas of Fig. 3) are usually poor immunogens. This has been found in a large set of experiments where native conserved HABPs were neither immunogenic nor protection-inducing when immunizing large numbers of Aotus monkeys. A different strategy had thus to be developed to render them immunogenic and protection inducing.
Some critical residues in RBC binding were replaced by glycine (the smallest amino acid and an α-helix breaker) in the first trials, trying to convert these conserved native HABPs' non-immunogenic and non-protection-inducing peptides into immunogenic and protection-inducing ones [82, 83].
Western blot with merozoite lysate determined their ability to induce antibodies against the P. falciparum malaria parasite as assessed by immunofluorescence (IFA) with air-dried, unfixed parasite cultures and native proteins. Their protection-inducing ability was tested when these monkeys were intravenously inoculated with a 100% infective Aotus-adapted strain of this parasite (FVO).
The phenomenon's fine-tuning, following hundreds of experiments performed on large numbers of Aotus monkeys [81–91] (Table 1), has allowed us to ascertain that some critical binding residues have to be replaced by others by specifically and reciprocally shifting their polarity but maintaining their other physico-chemical characteristics, such as mass, volume and surface. F could thus be replaced by R and vice versa; L↔H, P↔D, W↔Y, M↔K and E, I↔N, T↔C and V↔N or S. Serine, alanine and glycine remained undefined owing to their very small mass, similar polarity, small volume and the absence of counterparts of opposite polarity as occurs with other amino acids. A↔S sometimes worked, suggesting that some rules have still to be identified for these small residues [91] (Table 1).
Table 1.
An example of 4313, 4325 and 4337 (AMA-1 derived) HABP-induced humoral immune response and protective immunity and that of their modified analogues [87, 83, 88] Some of the amino acid replacements leading to the breaking of the immunological code of silence. Antibody titres were determined by immunofluorescence antibody assay (IFA) 15 days after the second and 15 days after the third immunization. Protection was defined as the complete absence of parasites in immunized monkey blood for 15 days following challenge. All control monkeys reached ≥6% parasitaemia by days 6–8; they were then treated, cured and released back into the Amazon jungle. ND, not determined. Modified HABP groups, A: immunogenic protection-inducer; B: long-lasting high antibody titre non-protection inducers; D: short-lived non-protection inducers; E: Non-immunogenic non-protection inducer. Boxed are the modified HABPs for which the 1H NMR structure has been shown.
 |
Table 2.
Amino acid sequences of some representative native and modified HABPs (in bold) derived from MSP-1 [81, 82, 84], EBA-175 [85], SERA [90], RESA-155 [89], MSP-2 [86] and AMA-1 [83, 87, 88] proteins. The letters a and b represent repetitions of these experiments in different groups of Aotus monkeys. Sequences have been aligned according to the correct identification of residues corresponding to modified HABP HLADR²1* binding motif and reading register in Pockets 1, 4, 6 and 9 [24, 91, 94]. Antibody titres were determined by IFA (shown in brackets) corresponding to sera dilution and the number of Aotus that developed such antibodies previous to challenge. Each group of monkeys received two or three doses of modified HABP. Prot. = the number of monkeys which were fully protected against intravenous inoculation of 100,000 infected erythrocytes from the P. falciparum Aotus adapted FVO strain. Parasitaemia was determined by the very sensitive Acridine Orange methodology.
 |
Such experiments with these immunogenic, protection-inducing, modified HABPs were repeated two or three times with new groups of Aotus monkeys, leading to similar results (Table 1, Group A experiments' b with the corresponding peptide) [91].
High antibody titres detected by IFA and Western blot were also induced by some other modified HABPs in some monkeys (Table 1, modified 14518, 14520, 17934, 17936, 4325 analogues in Group D) and detected 15 days after the second immunization. Such antibodies disappeared and did not reappear 15 days after the third dose, when their levels could be expected to increase following the third immunization. These peptides induced short-lived antibody responses [92] that did not induce protection against experimental infection.
Experimental challenge for Group B monkeys (shown in Table 1) revealed that some other modifications made to these conserved HABPs induced high long-lasting antibody titres as assessed by IFA and Western blot that did not lead to their inducing protection, since these Aotus became infected to the same extent as controls [93] (Table 1, Group B). However, it was found that these antibodies recognized different structures on the same molecule, but that only one specific modification was effective in inducing protective immunity. Both short-lived and long-lasting antibody-inducing modified HABP phenomena were explained at the atomic level by 1H-NMR structural analysis [92, 93].
Structural analysis of native and modified HABPs
When searching for an association between immunological activity and native and modified HABP structure as determined by H-NMR analysis it was noticed that the changes made induced striking structural modifications in these peptides (numbered according our Institute's code where bold is used throughout the rest of this article to distinguish modified HABPs from native ones shown in brackets). α-helixes were thus shortened in 24292 peptides derived from (1815), 23230 (6746), 13446 (1522) and 22812 (1779), α-helixes were displaced in 13450 (1585) and 20034 (4325), short α-helixes were induced in random-configuration peptides in 14044 (4337) and β-turn tendency modified as in 13492 (6671). Distorting classical-type III β-turns were induced as in 24112 (4044) or in 24166 (1818) (random structure that could not be classified by CD). All such modifications rendered these peptides immunogenic and protection-inducing against experimental challenge (Table 3).
Table 3.
Structural features and HLA-DR binding characteristics of some native and immunogenic, protection-inducing modified HABPs (in bold) included in this article Native and modified HABP secondary and tertiary structures determined by circular dichroism and 1H NMR, respectively. Inter-atomic distance is given in angstroms between the most distant hydrogen atoms from residues fitting into Pocket 1 to Pocket 9 from the corresponding HLA-DRβ1* alleles. The percentage of their binding to purified HLA-DRβ1* molecules (shaded) is grouped according to the corresponding haplotype. Putative TCR-contacting residues are also indicated.
| Peptide | Structural features | Dist.(A) | Haplotvpes | TCRcontact residues | Ref. | ||||
|---|---|---|---|---|---|---|---|---|---|
| DRll | DR52 | DR53 | |||||||
| %Binding to HLA-DRbi | |||||||||
| 0101 | 0301 | 1101 | 0401 | 0701 | |||||
| 1585 | α-helix Y4-Y12 | 14.3 | 5 | 18 | 15 | 7 | 37 | 82 | |
| 13450 | α-helix L5-Q18 | 20.2 | 12 | 47 | 20 | −8 | 33 | 3, 7, 8 | |
| 1815 | α-helix Q4-L14 | 17.7 | −2 | −10 | −10 | 30 | 45 | 91 | | |
| 24292 | α-helix I6-F12 | 19.6 | −4 | 51 | 42 | 16 | 27 | 3, 7, 8 | |
| 6746 | α-helix (CD) | - | 12 | 55 | 76 | 5 | 42 | 90 | |
| 23230 | α-helix N9-H16 | 20.0 | 2 | 84 | 37 | 13 | ND | 3, 7, 8 | |
| 1522 | α-helix Q1-V20 | 16.9 | 1 | 88 | 91 | −3 | 23 | 91 | |
| 13446 | α-helix P3-N11 | 20.2 | −3 | 43 | 77 | −3 | 25 | 3, 7, 8 | |
| 1585 | a-helix Y4-Y12 | 14.3 | 5 | 18 | 15 | 7 | 37 | 82 | |
| 10014 | a-helix A9-Q18 | 20.9 | −4 | 30 | 61 | −5 | 33 | 3, 7, 8 | |
| 1779 | a-helix (CD) | - | 12 | 55 | 76 | 5 | 31 | 109 | |
| 22812 | a-helix N9-H16 | 19.3 | 5 | 30 | 54 | 6 | ND | 3, 7, 8 | |
| 6671 | β-turn tendency (CD) | - | 24 | 45 | 16 | 61 | 55 | 89 | |
| 13492 | a-helix V4-R8 | 24.7 | −5 | 33 | 16 | 50 | 16 | 2, 3, 8 | |
| 4044 | Classical type III' β turn S7-F10 | 19.0 | 5 | 0 −60 | 38 | 39 | 86 | ||
| 24112 | Distorted type III' β turn Y3-T6and Classical type III β turn A11-M14 | 26.5 | 9 | 1 | 0 | 53 | 46 | 2, 3, 8 | |
| 4325 | β-turn and short α-helix K13-R16 | 21.5 | 18 | 83 | 74 | 28 | 48 | 83 | |
| 20034 | a-helix K3-F6 and A11-R16 | 25.5 | 40 | 34 | 7 | 69 | 64 | 2, 3, 8 | |
| 4313 | Random | - | 0 | 20 | 26 | 9 | 23 | 87 | |
| 10022 | Distorted type III' β turn T7-F10 | 25.0 | 9 | 7 | 4 | 2 | 42 | 2, 3, 8 | |
| 4337 | Random | - | −17 | 12 | −6 | 4 | 28 | 88 | |
| 14044 | short α-helix K5-T10 | 23.1 | 4 | 19 | 39 | 7 | 79 | 2, 3, 8 | |
| 1818 | Random | - | 2 | 33 | 6 | 11 | 36 | 3, 5, 8 | 91 |
| 24166 | Classical type III β turn S9-N12 and Distorted type III β turn Y14-M17 | 24.9 | 53 | 5 | 69 | 18 | 38 | ||
It was suggested that modifying these HABPs probably led to a better fit into HLA-DRβ1* molecules; their ability to bind to these MHC-II molecules was therefore analysed (Table 3).
This showed that native HABP activity (some did not bind to purified HLA-DRβ1* molecules) changed once they had been modified, since they bound with very high affinity and specificity to certain HLA-DR alleles, such as 13450, 24292 to HLA-DRβ1*0301, 10014 to HLA-DRβ1*1101, 24112 to HLA-DRβ1*0401, 10022 and 14044 to HLA-DRβ1*0701 and 24166 to HLA-DRβ1*0101 and 1101. Other promiscuous binding peptides, such as the 23230 (6746) analogue, modified their binding to specifically bind to HLA-DRβ1*0301, 13446 (derived from 1522) to HLA-DRβ1*1101, 13492 (6671) to HLA-DRp1*0401 and 22812 (1779) to HLA-DRp1*1101.
These tailored HABPs displayed binding motifs and reading registers specific for those alleles to which they bound [24, 94]. Peptides 13450, 24292 and 23230 thus bound to HLA-DRβ1*0301 having residues Y, I and W (respectively) fitting into this molecule's Pocket 1; D, D and A fitted into Pocket 4; A, E and K fitted into Pocket 6 and Y, L and L fitted into Pocket 9, all being specific amino acids conforming to this allele's HLA-DR^1*0301 binding motifs and reading registers. The same happened for other peptides binding to HLA-DRβ1*1101, HLA-DRβ1*0401, HLA-DRβ1*0701 and HLA-DR^1*0101 (Table 3).
Secondary structure analysis
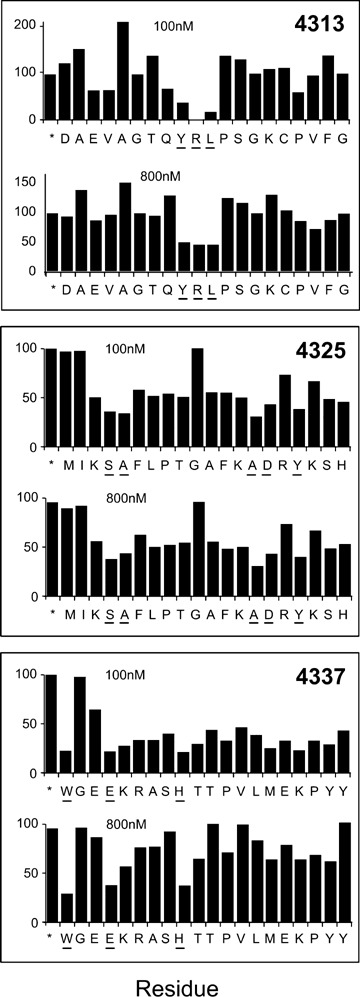
Native and modified HABP secondary structure circular dichroism (CD) analysis of 100 conserved malarial HABPs [95] identified in the 25 most relevant P. falciparum merozoite proteins involved in RBC invasion (Fig. 5), confirmed in most of them by 1H NMR, clearly showed these conserved HABPs' functional compartmen-talization. Those having an α-helical structure were associated with proteins or their fragments involved in invasion, which were processed or cleaved after fulfilling their function. The fragments where these conserved HABPs are included are released from the merozoite membrane and released into the milieu [96–98] after having mediated initial binding to RBCs.
Fig 5.
Secondary structural features for HABPs corresponding to 25 proteins involved in invasion. The figure shows the CD spectra of 94 HABPs [95] where α-helical structural elements were present in some HABPs according to 208 and 220 nm minimal values and 190 nm maximum elipcity, whereas the other HABPs displayed random coil structures having 200 nm minima and maxima below zero elipcity.
Those conserved HABPs present in proteins or their fragments remaining anchored to the membrane via a GPI tail, containing a Plasmodium exporting element (PEXEL) [99] or having a trans-membrane domain, which are the only ones present on newly infected RBCs, display a β-turn or random structure.
These findings clearly suggest an association between some structural elements present in conserved HABPs and their functional activity in the proteins involved in invasion [95]. They also suggest functional compartmentalization where α-helical HABPs are present in soluble fragments of proteins to be released into the milieu after performing their binding activities, while β-turn, or random HABPs, remain anchored to the merozoite's membrane and penetrate newly infected RBCs.
It was also found that most native HABPs presenting a predominantly β-helical structure specifically bound to HLA-DRβ1*0301 and 1101 alleles (belonging to the HLA-DR52 haplotype), that most native HABPs (predominantly displaying a β-turn structure) bound to HLA-DRβ1*0401 and that all native HABPs giving rise to modified, immunogenic and protective HABP binding to HLA-DRβ1*0701 mainly had random coil structure, the latter belonging to the HLA-DR53 haplotype. Native peptide 1818 (not classified by CD but adopting a β-turn structure when modified [24166]) bound to HLA-DRp1*0101 and HLA-DRp1*1101. These data also suggest immune system compartmentalization, where α-helical HABPs could preferentially bind to HLA-DR52 molecules, while the β-turn of random-structured conserved HABPs could bind to HLA-DR53 molecules, implying that native HABPs' secondary structure could suggest the modifications to be performed to fit into a specific HLA-DRβ1* allele or haplotype to render them immunogenic and protection-inducing (Fig. 5).
The Dictionary of Protein Secondary Structure (DPSS) [100] has identified three helix types (310 or G, a or H, π or I), turns (represented by T), strands (E), bridges (B), bends (S) and other secondary structures (L). Identifying such structures in conserved HABPs by an easy and accessible methodology such as CD analysis will lead to the easier design of immunogenic and protection-inducing peptides.
Native and modified HABP 3D structure explains some immunological phenomena
Given the critical importance of structural modifications performed on native HABPs for inducing protection, knowledge of these antigens' 3D structure is essential for completely understanding protection mechanisms.
1H NMR analysis of ∼100 of our modified and native HABP 3D structures revealed that all peptides binding to HLA-DRp1*0301 and HLA-DRβ1*1101 (HLA-DR52 haplotype members) had a 20 ± 1.5 Å distance between the farthest hydrogen from the residues fitting into these alleles' Pocket 1 (fuchsia) to Pocket 9 (green). All modified HABP binding to HLA-DRβ1*0401 and HLA-DRβ1*0701 (members of the HLA-DR53 haplotype) presented a 25 ± 1.5 Å distance between the same residues. The only modified HABP binding to the HLA-DRβ1*0101 allele (from haplotype DR1) had a 25 Å distance between residues fitting into the same pockets. These data clearly suggested an approximately 2-Å difference in distance between different HLA-DR haplotype molecules' Pockets 1 to 9 [91] (Fig. 6C).
Fig 6.
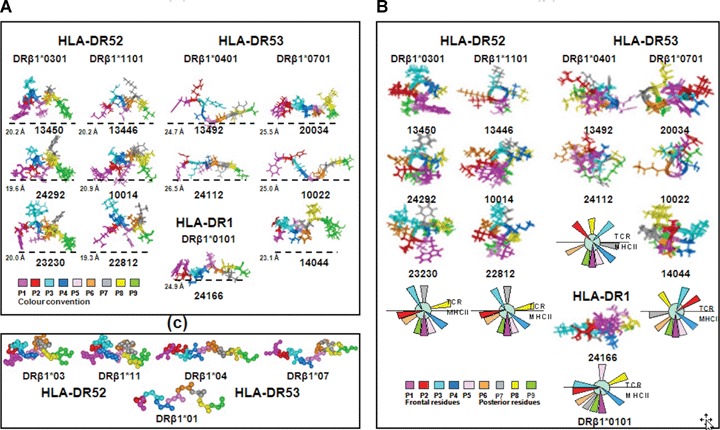
(A) Lateral view of the three-dimensional (3D) structure of some immunogenic protection-inducing modified malarial HABPs (determined by 1H NMR), according to their HLA-DRβ1* binding activity, binding motifs and reading registers. The only HLA-DRβ1*0101 peptide is shown at the bottom of the HLA-DRβ1*0401 column. Colour code for amino acids fitting into pockets and their localization is displayed at the bottom of each figure. Modified HABPs fitting into haplotype DR52 (HLA-DRβ1*0301 and HLA-DRβ1*1101) had 20 ± 1.5 Å distance and those fitting into purified HLA-DRβ1*0401 and HLA-DRβ1*0701 molecules (haplotype DR53) had a 25 ± 1.5 Å distance between the farthest atoms fitting into Pockets 1 to 9, supporting the existence of about 2 Å difference in distance between different HLA-DR haplotypes' Pockets 1 to 9. (B) Front view of some immuno-genic-protection-inducing peptides' 3D structure determined by 1H NMR. A consensus diagram of front-view residue orientation is shown at the bottom. Residues opposed and perpendicularly orientated downwards fit into P1 (fuchsia, forward) and P9 (green, backwards). Residues fitting into P4 (dark blue) and P6 (light brown) are orientated to adjust into the corresponding lateral pockets of the peptide-binding region shallow floor. Any amino acid orientated on the same plane (or lower down) suggested the existence of pockets other than 1, 4, 6 or 9 in these Class II molecules. A putative Pocket 2 for HLA-DR52 haplotype alleles and a putative Pocket 7 for HLA-DR53 haplotype molecules can thus be suggested since these residues are localized at the same HLA-DR platform level as canonical Pockets 4 and 6. Upwardly orientated residues, being opposite to these pockets, could be binding to the TCR; therefore, residues 3, 7 and 8 in HLA-DR52 alleles 2, 3 and 8 in HLA- DR53 alleles and residues 3, 5 and 8 for haplotype DR1 could be making contact with the TCR. All these data suggest haplotype- and allele-conscious TCR differences in immunogenic-protection-inducing anti-malarial modified HABPs. (C) Top view of the backbone atoms of the superimposed immunogenic, protection-inducing, modified, malarial HABPs binding to the HLA-DR molecules displayed above showing the different distances between these residues fitting into Pockets 1 (fuchsia) and 9 (green), among the different haplotype-binding HABPs and their different orientation between residues P2 (red) and P7 (grey) amongst different haplotypes and alleles binding modified HABPs. RMSD was <1.00. Such differences in length and orientation for the different haplotypes have to be seriously taken into account when designing vaccines.
1H NMR analysis has also revealed that orientation varied in modified HABP residues according to the haplotypes to which they bound (Fig. 6A and B), such as residue P2 (red) horizontally or downwardly orientated from the plane formed by Pockets 4 (dark blue) and 6 (light brown) in modified HABP binding to HLA-DRp1*0301 and 1101 (HLA-DR52). Such orientation suggested a putative Pocket 2 in HLA-DR52 alleles while this P2 residue (red) was fully upwardly orientated in modified HABP binding to HLA-DRβ1*0401 and 0701 (towards the left of the frontal plane initiated by P1 in the HLA-DRβ1*0401 and towards the right in HLA-DRβ1*0701 on the same plane), suggesting that they could be making contact with the TCR and have different orientation in these two alleles from the same HLA-DR53 haplotype (Fig. 6A and B).
Similar behaviour could be observed (Fig. 6A and B) for residue P7 (grey), horizontally or downwardly orientated in haplotype DR53 alleles (HLA-DRβ1*0401 and 0701), again towards the right in HLA-DRβ1*0401 and the left in HLA-DRβ1*0701 on the same plane, suggesting a probable Pocket 7 having different lateral orientations in these two alleles. However, it was upwardly orientated in haplotype DR52 alleles (HLA-DRβ1*0301 and 1101), suggesting P7 contact with the TCR.
Amino acid residues theoretically made contact with the TCR in modified HABP binding to HLA-DRβ1*0301 and HLA-DRβ1*1101 in P3 (aquamarine), P7 (grey) and P8 (yellow). Those residues making contact with the TCR in modified HABP binding to HLA-DRβ1*0401 and HLA-DRp1*0701 did so in P2 (red), P3 (aquamarine) and P8 (yellow) (Fig. 7E), whilst modified HABP residues bound to HLA-DR_1*0101 (from haplotype DR1) in P3 (aquamarine), P5 (pink) and P8 (yellow) (Fig. 6A and B; Table 3). Identical results have been found for many other peptides [81–91].
Fig 7.
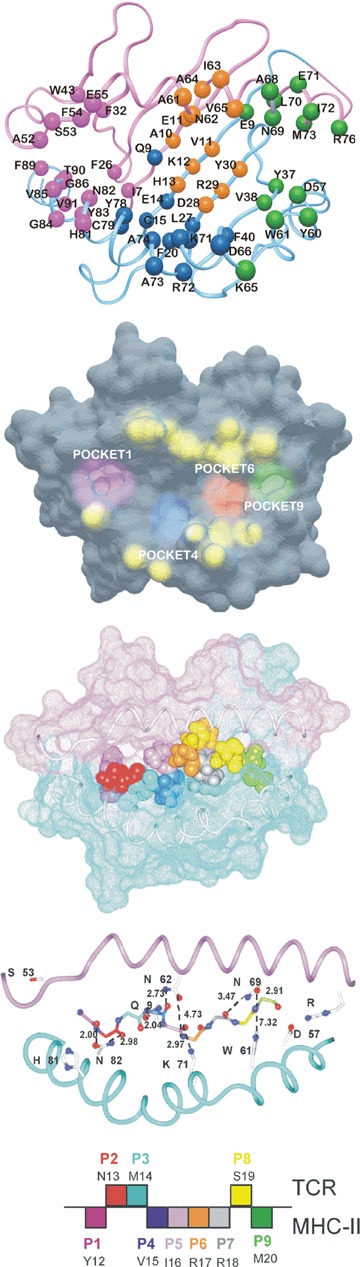
(A) Ribbon representation of the HLA-DRβ1*0401 [103] (PDB accession number 2SEB) molecule showing the A chain in pink and the β chain in light blue. Localization of residues forming Pocket 1 are shown in fuchsia, Pocket 4 in blue, Pocket 6 in light brown and Pocket 9 in green. (B) HLA-DRβ1*0401 surface [103], showing the deep localization of Pockets 1 (fuchsia) and Pocket 9 (green) and the superficial localization of Pockets 4 (dark blue) and 6 (light brown) on the PBR platform of this HLA-DRβ1*0401 molecule. The TCR contacting residues on this molecule are displayed in yellow and show the elevated prominence above this molecule's peptide-binding residue (PBR) groove. (C) Docking and molecular modelling of malarial immunogenic, protection-inducing, modified HABP 24112 residues (12YNMVIRRSM20) fitting into the HLA-DRβ1*0401 molecule's PBR based on this peptide's binding activity, binding motifs and reading registers for this Class II molecule. Van der Waals surface amino acid colour code is displayed at the bottom of this figure in panel c. Note the fit of residues Y12 (fuchsia), V15 (dark blue), R17 (light brown) and M20 (green) into the corresponding pockets P1, P4, P6 and P9, including R18 (grey) in P7, confirming the suggested existence of this pocket in the HLA-DRβ1*0401 molecule. (D) This shows the H bonds spontaneously established between amino acids Qα9, Nα62, Nα69, Nβ82, Kβ71 and Wβ61 of HLA-DRβ1*0401 (code 2SEB in PDB) [103] and the backbone of modified HABP 24112. Seven out of nine residues showed a distance <3.5 Å, characteristic of the H bonds with which the peptide is anchored into the PBR of these Class II molecules to conform a stable MHCII–peptide–TCR complex. It has also to be stressed that both structures were determined by very different methods: X-ray crystallography for HLA-DRβ1*0401 and 1H NMR in solution for HABP 24112. In spite of these methodological differences, RMSD between Col II peptide with which HLA-DRβ1*0401 was crystallized and modified HABP 24112 was only 1.82 Å. (E) Diagrammatic representation of 24112 residues orientation fitting into HLA-DRβ1*0401 pockets (downwardly oriented) and TCR (upwardly oriented). Colour code as previously described.
Supporting the haplotype- and allele-conscious TCR concept
The above findings were supported by X-ray crystallography studies of HLA-DRβ1*0401 molecule binding haemagglutinin (HA) or collagen (Col II) peptide where it was found that the bound peptide's amino acid sequence determined the width of the HLA-DRp1*0401 binding groove. It was narrower in the HLA-DRβ1*0401-Col II complex by 2.0 Å, suggesting peptide binding region (PBR) mobility in any direction [101]. This article shows an ∼2.0 Å difference between Pockets 1 and 9 amongst different haplotypes. A shallow, hydrophobic Pocket 7 was also clearly identified in the HLA-DRβ1*0401-Col II complex. A Pocket 7, for which specific binding amino acids have been identified as specific binding motifs, has also been described for HLA-DRβ1*04 molecules [24, 94]. This strongly supports our findings concerning the existence of some other pockets (different to the canonical ones) characteristic for the different haplotypes and alleles.
Functional and structural differences could therefore have occurred since the HLA-DRβ1*04 allelic lineage (DR53) has been estimated to be 85 million years old, while the HLA-DR52 haplotype originated from two successive gene duplication events estimated to have occurred 60 and 40 million years ago [102].
Modified HABPs' 3D structure revealed a fit into HLA molecules
The docking and molecular modelling of modified HABP 24112 (immunogenic and protection-inducing) into the HLA-DRβ1*0401 molecule (code 2SEB in PDB) [103] (Fig. 7A and B) to which it binds with high affinity, provides an example of a tailor-made, modified, conserved malarial HABP's perfect fit. HABP 24112 presented an optimum fit into this molecule's peptide-binding region (Fig. 7C), spontaneously forming seven of the nine H-bonds between the peptide backbone and the HLA-DRβ1*0401 residues anchoring the peptide to such molecule. HABP 24112 backbone presented 1.86 RMSD when it was superimposed on the Col II peptide backbone [103].
Figure 7D shows the spontaneously formed H-bonds between O81 from Asn 82β with the peptide's N from N13 (atoms are underlined in the peptide for clarifying interactions), Nδ2 from Asn82β with O from N13, Oɛ1 from Gln 9a with N from V15, Oδ1 from Asn62α with O from V15, Nz from Lys 71 β with O from I16, Oδ1 from Asn 69α with N from M20 and N82 from Asn69a with O from
R18. These H-bonds between the peptide and the lateral chains from the previously mentioned amino acids stabilized peptide binding to these Class II molecules; (>3.5 Å distances were manually established only between O81 from Asn 62a with N from Arg 17 and Nɛ1 from Trp 61 β with O from S19 from HABP 24112 backbone.
H-bond formation between αS53 and βH81 was not analysed as residues -P1 and -P2 were outside HLA-DRβ1*0401 pockets. These two completed the 11 H bonds established between these amino acids and the peptide backbone stabilizing peptide binding to MHC II molecules. It should be stressed that Aotus MHC-DRB*W47 allele (for which 24112 is highly specific) exhibited the highest Class II molecular homology (97–100%) with human HLA-DRβ1*0403/0407 [28].
Varied crossing angle geometry in the MHC-Class II–peptide-TCR complexes, ranging from 40° to 87°, from diagonal [104] to near orthogonal [105] and some additional lateral mobility along the groove [106] that can displace some TCRs from their roughly central location towards either end of the binding groove, led us to propose the existence of haplotype- and allele-conscious TCR as seen in modified HABP 24112.
Conclusion
Modifying some critical binding residues in these HABP attachments to RBCs allowed them to induce immunogenicity and protection against experimental challenge, probably as a consequence of having acquired the ability to bind to specific HLA-DRβ1* alleles.
These modified HABPs, displaying characteristic structural patterns in their native form, suggest functional compartmental-ization associated with their localization in the invasion-mediating protein fragment, where those present in cleaved and released fragments mainly display an α-helical structure. Those anchored to the membrane via a GPI tail, having a transmembranal domain or PEXEL-associated motif, displayed β-turn or random structural patterns.
By the same token, those displaying α-helical structures mainly bound to HLA-DR52 and DR8 haplotype alleles while those having β-turn or random structure mainly bound to HLA-DR53 and DR1 molecules, also suggesting these conserved HABPs' immunological compartmentalization-associated 3D structure.
These HABPs also adopted certain characteristic structural configurations associated with their HLA-DR haplotype binding activities and characteristics, such as a 2 Å distance difference between amino acids fitting into HLA-DRp1* HLA-DR52, DR8 and HLA-DR53 DR1 Pockets 1 to 9, suggesting haplotype- and allele-conscious TCR. Vaccine development must thus take the epitopes' structural characteristics and the host's immunogenetic characteristics into account.
Our quantum chemistry studies have led us to ascertain that differences observed in amino acid sequences between human and Aotus Class II molecules [107, 108] are not relevant, since very few of these changes involved variations in these molecules' pockets.
Acknowledgments
This research has been supported by COLCIENCIAS contract RC-41 2007. The collaboration of our chemistry and immunology sections is greatly appreciated, as is that of the whole staff at FIDIC. We would especially like to thank Jason Garry for patiently translating and reading the manuscript.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–7. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–8. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 4.Florens L, Liu X, Wang Y, Yang S, Schwartz O, Peglar M, Carucci DJ, Yates JR, 3rd, Wub Y. Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol Biochem Parasitol. 2004;135:1–11. doi: 10.1016/j.molbiopara.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez LE, Curtidor H, Valbuena J, Puentes A, Vera-Bravo R, Urquiza M, Cifuentes G, Reyes C, Patarroyo M. Intimate molecular interactions of P. falci-parum merozoite proteins involved in invasion of red blood cells and their implications in vaccine design. Chem Rev. 2007 doi: 10.1021/cr068407v. ; submitted. [DOI] [PubMed] [Google Scholar]
- 6.Stubbs J, Simpson KM, Triglia T, Plouffe D, Tonkin CJ, Duraisingh MT, Maier AG, Winzeler EA, Cowman AF. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science. 2005;309:1384–7. doi: 10.1126/science.1115257. [DOI] [PubMed] [Google Scholar]
- 7.Coley AM, Parisi K, Masciantonio R, Hoeck J, Casey JL, Murphy VJ, Harris KS, Batchelor AH, Anders RF, Foley M. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect Immun. 2006;74:2628–36. doi: 10.1128/IAI.74.5.2628-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carruthers VB, Blackman MJ. A new release on life: emerging concepts in pro-teolysis and parasite invasion. Mol Microbiol. 2005;55:1617–30. doi: 10.1111/j.1365-2958.2005.04483.x. [DOI] [PubMed] [Google Scholar]
- 9.Kauth CW, Woehlbier U, Kern M, Mekonnen Z, Lutz R, Mucke N, Langowski J, Bujard H. Interactions between merozoite surface proteins 1, 6, and 7 of the malaria parasite Plasmodium falciparum. J Biol Chem. 2006;281:31517–27. doi: 10.1074/jbc.M604641200. [DOI] [PubMed] [Google Scholar]
- 10.Garcia JE, Puentes A, Patarroyo ME. Developmental biology of sporozoite-host interactions in Plasmodium falciparum malaria: implications for vaccine design. Clin Microbiol Rev. 2006;19:686–707. doi: 10.1128/CMR.00063-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patarroyo ME, Romero P, Torres ML, Clavijo P, Moreno A, Martinez A, Rodriguez R, Guzman F, Cabezas E. Induction of protective immunity against experimental infection with malaria using synthetic peptides. Nature. 1987;328:629–22. doi: 10.1038/328629a0. [DOI] [PubMed] [Google Scholar]
- 12.Patarroyo ME, Amador R, Clavijo P, Moreno A, Guzman F, Romero P, Tascon R, Franco A, Murillo LA, Ponton G, Trujillo G. A synthetic vaccine protects humans against challenge with asexual blood stages of Plasmodium falciparum malaria. Nature. 1988;332:158–61. doi: 10.1038/332158a0. [DOI] [PubMed] [Google Scholar]
- 13.Valero MV, Amador LR, Galindo C, Figueroa J, Bello MS, Murillo LA, Mora AL, Patarroyo G, Rocha CL, Rojas M, Aponte JJ, Sarmiento LE, Losada DM, Coronell CG, Ortega NN, Rosas JE, Alonso PL, Pataroyo ME. Vaccination with SPf66, a chemically synthesised vaccine, against Plasmodium falciparum malaria in Colombia. Lancet. 1993;341:705–10. doi: 10.1016/0140-6736(93)90483-w. [DOI] [PubMed] [Google Scholar]
- 14.Noya O, Gabaldon Berti Y, Alarcon de, Noya B, Borges R, Zerpa N, Urbaez JD, Madonna A, Garrido E, Jimenez MA, Borges RE, Garcia P, Reyes I, Prieto W, Colmenares C, Pabon R, Barraez T, Caceres LG, Godoy N, Cifontes R. A population-based clinical trial with the SPf66 synthetic Plasmodium falciparum malaria vaccine in Venezuela. J Infect Dis. 1994;170:396–402. doi: 10.1093/infdis/170.2.396. [DOI] [PubMed] [Google Scholar]
- 15.Sempertegui F, Estrella B, Moscoso J, Piedrahita L, Hernandez D, Gaybor J, Naranjo P, Mancero O, Arias S, Bernal R, Cordova ME, Suarez J, Zicker F. Safety, immunogenicity and protective effect of the SPf66 malaria synthetic vaccine against Plasmodium falciparum infection in a randomized double-blind placebo-controlled field trial in an endemic area of Ecuador. Vaccine. 1994;12:337–42. doi: 10.1016/0264-410x(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 16.Alonso PL, Smith T, Schellenberg JR, Masanja H, Mwankusye S, Urassa H, Bastos de Azevedo I, Chongela J, Kobero S, Menendez C, Hurt N, Thomas MC, Lyimo E, Weiss NA, Hayes R, Kitua AY, Lopez MC, Kilama WL. Teuscher T, Taner M. Randomised trial of efficacy of SPf66 vaccine against Plasmodium falciparum malaria in children in southern Tanzania. Lancet. 1994;344:1175–81. doi: 10.1016/s0140-6736(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 17.Valero MV, Amador R, Aponte JJ, Narvaez A, Galindo C, Silva Y, Rosas J, Guzman F, Patarroyo ME. Evaluation of SPf66 malaria vaccine during a 22-month follow-up field trial in the Pacific coast of Colombia. Vaccine. 1996;14:1466–70. doi: 10.1016/s0264-410x(96)00070-9. [DOI] [PubMed] [Google Scholar]
- 18.Acosta CJ, Galindo CM, Schellenberg D, Aponte JJ, Kahigwa E, Urassa H, Schellenberg JR, Masanja H, Hayes R, Kitua AY, Lwilla F, Mshinda H, Menendez C, Tanner M, Alonso PL. Evaluation of the SPf66 vaccine for malaria control when delivered through the EPI scheme in Tanzania. Trop Med Int Health. 1999;4:368–96. doi: 10.1046/j.1365-3156.1999.00406.x. [DOI] [PubMed] [Google Scholar]
- 19.Nosten F, Luxemburger C, Kyle DE, Ballou WR, Wittes J, Wah E, Chongsuphajaisiddhi T, Gordon DM, White NJ, Sadoff JC, Heppner DG. Randomised double-blind placebo-controlled trial of SPf66 malaria vaccine in children in northwestern Thailand Shoklo SPf66 Malaria Vaccine Trial Group. Lancet. 1996;348:701–7. doi: 10.1016/s0140-6736(96)04465-0. [DOI] [PubMed] [Google Scholar]
- 20.Graves P, Gelband H. Vaccines for preventing malaria (Blood stage) Cochrane Database Syst Rev. 2006;4:CD006199. doi: 10.1002/14651858.CD006199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calvo M, Guzman F, Perez E, Segura CH, Molano A, Patarroyo ME. Specific interactions of synthetic peptides derived from P. falciparummerozoite proteins with human red blood cells. Pept Res. 1991;4:324–33. [PubMed] [Google Scholar]
- 22.Patarroyo ME, Vinasco J, Amador R, Espejo F, Silva Y, Moreno A, Rojas M, Mora AL, Salcedo M, Valero V, Goldberg AK, Kalil J. Genetic control of the immune response to a synthetic vaccine against Plasmodium falciparum. Parasite Immunol. 1991;13:509–16. doi: 10.1111/j.1365-3024.1991.tb00547.x. [DOI] [PubMed] [Google Scholar]
- 23.Murillo LA, Rocha CL, Mora AL, Kalil J, Goldenberg AK, Patarroyo ME. Molecular analysis of HLA DR4-beta 1 gene in malaria vaccinees. Typing and subtyping by PCR technique and oligonucleotides. Parasite Immunol. 1991;13:201–10. doi: 10.1111/j.1365-3024.1991.tb00275.x. [DOI] [PubMed] [Google Scholar]
- 24.Marsh SGP, Barber LD. The HLA facts book. Academic Press; 2000. New York: [Google Scholar]
- 25.Andersson G, Andersson L, Larhammar D, Rask L, Sigurdardottir S. Simplifying genetic locus assignment of HLA-DRB genes. Immunol Today. 1994;15:58–62. doi: 10.1016/0167-5699(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 26.Pico de Coana Y, Rodríguez J, Guerrero E, Barrero C, Rodriguez R, Mendoza M, Patarroyo MA. A highly infective Plasmodium vivaxstrain adapted to Aotus monkeys: quantitative haematological and molecular determinations useful for P. vivax malaria vaccine development. Vaccine. 2003;21:3930–7. doi: 10.1016/s0264-410x(03)00278-0. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez R, Moreno A, Guzman F, Calvo M, Patarroyo ME. Studies in owl monkeys leading to the development of a synthetic vaccine against the asexual blood stages of Plasmodium falciparum. Am J Trop Med Hyg. 1990;43:339–54. doi: 10.4269/ajtmh.1990.43.339. [DOI] [PubMed] [Google Scholar]
- 28.Suarez CF, Patarroyo ME, Trujillo E, Estupinan M, Baquero JE, Parra C, Rodriguez R. Owl monkey MHC-DRB exon 2 reveals high similarity with several HLA-DRB lineages. Immunogenetics. 2006;58:542–58. doi: 10.1007/s00251-006-0127-0. [DOI] [PubMed] [Google Scholar]
- 29.Moncada CA, Guerrero E, Cardenas P, Suarez CF, Patarroyo ME, Patarroyo MA. The T-cell receptor in primates: identifying and sequencing new owl monkey TRBV gene sub-groups. Immunogenetics. 2005;57:42–52. doi: 10.1007/s00251-004-0758-y. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez EC, Suarez CF, Mendez JA, Echeverry SJ, Murillo LA, Patarroyo ME. Identification, cloning, and sequencing of different cytokine genes in four species of owl monkey. Immunogenetics. 2002;54:645–53. doi: 10.1007/s00251-002-0512-2. [DOI] [PubMed] [Google Scholar]
- 31.Day M. Experts back research using primates. Br Med J. 2006;333:1235. doi: 10.1136/bmj.39063.532917.DB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–9. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 33.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–66. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Chitnis CE. Molecular insights into receptors used by malaria parasites for erythrocyte invasion. Curr Opin Hematol. 2001;8:85–91. doi: 10.1097/00062752-200103000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Rener J, Graves PM, Carter R, Williams JL, Burkot TR. Target antigens of transmission-blocking immunity on gametes of Plasmodium falciparum. J Exp Med. 1983;158:976–81. doi: 10.1084/jem.158.3.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holder AA, Lockyer MJ, Odink KG, Sandhu JS, Riveros-Moreno V, Nicholls SC, Hillman Y, Davey LS, Tizard ML, Schwarz RT, Freeman RR. Primary structure of the precursor to the three major surface antigens of Plasmodium falciparum merozoites. Nature. 1985;317:270–3. doi: 10.1038/317270a0. [DOI] [PubMed] [Google Scholar]
- 37.Marshall VM, Silva A, Foley M, Cranmer S, Wang L, McColl DJ, Kemp DJ, Coppel RL. A second merozoite surface protein (MSP-4) of Plasmodium falciparum that contains an epidermal growth factor-like domain. Infect Immun. 1997;65:4460–7. doi: 10.1128/iai.65.11.4460-4467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Black CG, Wu T, Wang L, Hibbs AR, Coppel RL. Merozoite surface protein 8 of Plasmodium falciparum contains two epidermal growth factor-like domains. Mol Biochem Parasitol. 2001;114:217–26. doi: 10.1016/s0166-6851(01)00265-1. [DOI] [PubMed] [Google Scholar]
- 39.Gerold P, Schofield L, Blackman MJ, Holder AA, Schwarz RT. Structural analysis of the glycosyl-phosphatidylinositol membrane anchor of the merozoite surface proteins-1 and -2 of Plasmodium falciparum. Mol Biochem Parasitol. 1996;75:131–43. doi: 10.1016/0166-6851(95)02518-9. [DOI] [PubMed] [Google Scholar]
- 40.Weber JL, Lyon JA, Wolff RH, Hall T, Lowell GH, Chulay JD. Primary structure of a Plasmodium falciparum malaria antigen located at the merozoite surface and within the parasitophorous vacuole. J Biol Chem. 1988;263:11421–5. [PubMed] [Google Scholar]
- 41.McColl DJ, Anders RF. Conservation of structural motifs and antigenic diversity in the Plasmodium falciparum merozoite surface protein-3 (MSP-3) Mol Biochem Parasitol. 1997;90:21–31. doi: 10.1016/s0166-6851(97)00130-8. [DOI] [PubMed] [Google Scholar]
- 42.Pachebat JA, Ling IT, Grainger M, Trucco C, Howell S, Fernandez-Reyes D, Gunaratne R, Holder AA. The 22 kDa component of the protein complex on the surface of Plasmodium falciparum merozoites is derived from a larger precursor, merozoite surface protein 7. Mol Biochem Parasitol. 2001;117:83–9. doi: 10.1016/s0166-6851(01)00336-x. [DOI] [PubMed] [Google Scholar]
- 43.Trucco C, Fernandez-Reyes D, Howell S, Stafford WH, Scott-Finnigan TJ, Grainger M, Ogun SA, Taylor WR, Holder AA. The merozoite surface protein 6 gene codes for a 36 kDa protein associated with the Plasmodium falciparum merozoite surface protein-1 complex. Mol Biochem Parasitol. 2001;112:91–101. doi: 10.1016/s0166-6851(00)00350-9. [DOI] [PubMed] [Google Scholar]
- 44.LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR, Schoenfeld LW, Ota I, Sahasrabudhe S, Kurschner C, Fields S, Hughes RE. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–7. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 45.Black CG, Wang L, Wu T, Coppel RL. Apical location of a novel EGF-like domain-containing protein of Plasmodium falciparum. Mol Biochem Parasitol. 2003;127:59–68. doi: 10.1016/s0166-6851(02)00308-0. [DOI] [PubMed] [Google Scholar]
- 46.Adams JH, Blair PL, Kaneko O, Peterson DS. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 2001;17:297–9. doi: 10.1016/s1471-4922(01)01948-1. [DOI] [PubMed] [Google Scholar]
- 47.Sim BK, Orlandi PA, Haynes JD, Klotz FW, Carter JM, Camus D, Zegans ME, Chulay JD. Primary structure of the 175K Plasmodium falciparum erythrocyte binding antigen and identification of a peptide which elicits antibodies that inhibit malaria merozoite invasion. J Cell Biol. 1990;111:1877–84. doi: 10.1083/jcb.111.5.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson JK, Triglia T, Reed MB, Cowman AF. A novel ligand from Plasmodium falciparum that binds to a sialic acid-containing receptor on the surface of human erythrocytes. Mol Microbiol. 2001;41:47–58. doi: 10.1046/j.1365-2958.2001.02484.x. [DOI] [PubMed] [Google Scholar]
- 49.Gilberger TW, Thompson JK, Triglia T, Good RT, Duraisingh MT, Cowman AF. A novel erythrocyte binding antigen-175 par-alogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J Biol Chem. 2003;278:14480–6. doi: 10.1074/jbc.M211446200. [DOI] [PubMed] [Google Scholar]
- 50.Adams JH, Sim BK, Dolan SA, Fang X, Kaslow DC, Miller LH. A family of erythrocyte binding proteins of malaria parasites. Proc Natl Acad Sci USA. 1992;89:7085–9. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rayner JC, Vargas-Serrato E, Huber CS, Galinski MR, Barnwell JW. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resist-ant erythrocyte invasion pathway. J Exp Med. 2001;194:1571–81. doi: 10.1084/jem.194.11.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baldi DL, Good R, Duraisingh MT, Crabb BS, Cowman AF. Identification and disruption of the gene encoding the third member of the low-molecular-mass rhoptry complex in Plasmodium falciparum. Infect Immun. 2002;70:5236–45. doi: 10.1128/IAI.70.9.5236-5245.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Mohandas N, Thomas A, Coppel RL. Detection of detergent-resistant membranes in asexual blood-stage parasites of Plasmodium falciparum. Mol Biochem Parasitol. 2003;130:149–53. doi: 10.1016/s0166-6851(03)00165-8. [DOI] [PubMed] [Google Scholar]
- 54.Sanders PR, Gilson PR, Cantin GT, Greenbaum DC, Nebl T, Carucci DJ, McConville MJ, Schofield L, Hodder AN, Yates JR, 3rd, Crabb BS. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J Biol Chem. 2005;280:40169–76. doi: 10.1074/jbc.M509631200. [DOI] [PubMed] [Google Scholar]
- 55.Wang HY, Tang H, Shen CK, Wu CI. Rapidly evolving genes in human. I. The glycophorins and their possible role in evading malaria parasites. Mol Biol Evol. 2003;20:1795–804. doi: 10.1093/molbev/msg185. [DOI] [PubMed] [Google Scholar]
- 56.Duraisingh MT, Triglia T, Ralph SA, Rayner JC, Barnwell JW, McFadden GI, Cowman AF. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 2003;22:1047–57. doi: 10.1093/emboj/cdg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duraisingh MT, Maier AG, Triglia T, Cowman AF. Erythrocyte-binding antigen 175 mediates invasion in Plasmodium falciparum utilizing sialic acid-dependent and-independent pathways. Proc Natl Acad Sci USA. 2003;100:4796–801. doi: 10.1073/pnas.0730883100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goel VK, Li X, Chen H, Liu SC, Chishti AH, Oh SS. Band 3 is a host receptor binding merozoite surface protein 1 during the Plasmodium falciparum invasion of erythrocytes. Proc Natl Acad Sci USA. 2003;100:5164–9. doi: 10.1073/pnas.0834959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baum J, Maier AG, Good RT, Simpson KM, Cowman AF. Invasion by P. falciparum merozoites suggests a hierarchy of molecular interactions. PLoS Pathog. 2005;1:e37. doi: 10.1371/journal.ppat.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engers HD, Godal T. Malaria vaccine development: current status. Parasitol Today. 1998;14:56–64. doi: 10.1016/s0169-4758(97)01184-8. [DOI] [PubMed] [Google Scholar]
- 61.Graves P, Gelband H. Vaccines for preventing malaria. The Cochrane Library. 2007;(Issue 3) [Google Scholar]
- 62.Moorthy VS, Imoukhuede EB, Milligan P, Bojang K, Keating S, Kaye P, Pinder M, Gilbert SC, Walraven G, Greenwood BM, Hill AS. A randomised, double-blind, controlled vaccine efficacy trial of DNA/MVA ME-TRAP against malaria infection in Gambian adults. PLoS Med. 2004;1:e33. doi: 10.1371/journal.pmed.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bejon P, Mwacharo J, Kai O, Mwangi T, Milligan P, Todryk S, Keating S, Lang T, Lowe B, Gikonyo C, Molyneux C, Fegan G, Gilbert SC, Peshu N, Marsh K, Hill AV. A phase 2b randomised trial of the candidate malaria vaccines FP9 ME-TRAP and MVA ME-TRAP among children in Kenya. PLoS Clin Trials. 2006;1:e29. doi: 10.1371/journal.pctr.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sedegah M, Charoenvit Y, Minh L, Belmonte M, Majam VF, Abot S, Ganeshan H, Kumar S, Bacon DJ, Stowers A, Narum DL, Carucci DJ, Rogers WO. Reduced immunogenicity of DNA vaccine plasmids in mixtures. Gene Ther. 2004;11:448–56. doi: 10.1038/sj.gt.3302139. [DOI] [PubMed] [Google Scholar]
- 65.Alonso PL, Sacarlal J, Aponte JJ, Leach A, Macete E, Aide P, Sigauque B, Milman J, Mandomando I, Bassat Q, Guinovart C, Espasa M, Corachan S, Lievens M, Navia MM, Dubois MC, Menendez C, Dubovsky F, Cohen J, Thompson R, Ballou WR. Duration of protection with RTS,S/AS02A malaria vaccine in prevention of Plasmodium falciparum disease in Mozambican children: single-blind extended follow-up of a randomised controlled trial. Lancet. 2005;366:2012–8. doi: 10.1016/S0140-6736(05)67669-6. [DOI] [PubMed] [Google Scholar]
- 66.Pombo DJ, Lawrence G, Hirunpetcharat C, Rzepczyk C, Bryden M, Cloonan N, Anderson K, Mahakunkijcharoen Y, Martin LB, Wilson D, Elliott S, Elliott S, Eisen DP, Weinberg JB, Saul A, Good MF. Immunity to malaria after administration of ultra-low doses of red cells infected with Plasmodium falciparum. Lancet. 2002;360:610–7. doi: 10.1016/S0140-6736(02)09784-2. [DOI] [PubMed] [Google Scholar]
- 67.Genton B, Al-Yaman F, Betuela I, Anders RF, Saul A, Baea K, Mellombo M, Taraika J, Brown GV, Pye D, Irving DO, Felger I, Beck HP, Smith TA, Alpers MP. Safety and immunogenicity of a three-component blood-stage malaria vaccine (MSP1, MSP2, RESA) against Plasmodium falciparum in Papua New Guinean children. Vaccine. 2003;22:30–41. doi: 10.1016/s0264-410x(03)00536-x. [DOI] [PubMed] [Google Scholar]
- 68.Ockenhouse CF, Sun PF, Lanar DE, Wellde BT, Hall BT, Kester K, Stoute JA, Magill A, Krzych U, Farley L, Wirtz RA, Sadoff JC, Kaslow DC, Kumar S, Church LW, Crutcher JM, Wizel B, Hoffman S, Lalvani A, Hill AV, Tine JA, Guito KP, de Taisne C, Anders R, Horii T, Paolletti E, Ballou WR. Phase I/IIa safety, immunogenicity, and efficacy trial of NYVAC-Pf7, a pox-vectored, multiantigen, multistage vaccine candidate for Plasmodium falciparum malaria. J Infect Dis. 1998;177:1664–73. doi: 10.1086/515331. [DOI] [PubMed] [Google Scholar]
- 69.Genton B, Betuela I, Felger I, Al-Yaman F, Anders RF, Saul A, Rare L, Baisor M, Lorry K, Brown GV, Pye D, Irving DO, Smith TA, Beck HP, Alpers MP. A recom-binant blood-stage malaria vaccine reduces Plasmodium falciparum density and exerts selective pressure on parasite populations in a phase 1–2b trial in Papua New Guinea. J Infect Dis. 2002;185:820–7. doi: 10.1086/339342. [DOI] [PubMed] [Google Scholar]
- 70.Urquiza M, Rodriguez LE, Suarez JE, Guzman F, Ocampo M, Curtidor H, Segura C, Trujillo E, Patarroyo ME. Identification of Plasmodium falciparum MSP-1 peptides able to bind to human red blood cells. Parasite Immunol. 1996;18:515–26. doi: 10.1046/j.1365-3024.1996.d01-15.x. [DOI] [PubMed] [Google Scholar]
- 71.Rodriguez LE, Urquiza M, Ocampo M, Suarez J, Curtidor H, Guzman F, Vargas LE, Trivinos M, Rosas M, Patarroyo ME. Plasmodium falciparum EBA-175 kDa protein peptides which bind to human red blood cells. Parasitology. 2000;120:225–35. doi: 10.1017/s003118209900551x. [DOI] [PubMed] [Google Scholar]
- 72.Vera Bravo R, Marin V, Garcia J, Urquiza M, Torres E, Trujillo M, Rosas J, Patarroyo ME. Amino terminal peptides of the ring infected erythrocyte surface antigen of Plasmodium falciparum bind specifically to erythrocytes. Vaccine. 2000;18:1289–93. doi: 10.1016/s0264-410x(99)00405-3. [DOI] [PubMed] [Google Scholar]
- 73.Ocampo M, Urquiza M, Guzman F, Rodriguez LE, Suarez J, Curtidor H, Rosas J, Diaz M, Patarroyo ME. Two MSA 2 peptides that bind to human red blood cells are relevant to Plasmodium falciparum merozoite invasion. J Pept Res. 2000;55:216–23. doi: 10.1034/j.1399-3011.2000.00174.x. [DOI] [PubMed] [Google Scholar]
- 74.Urquiza M, Suarez JE, Cardenas C, Lopez R, Puentes A, Chavez F, Calvo JC, Patarroyo ME. Plasmodium falciparum AMA-1 erythrocyte binding peptides implicate AMA-1 as erythrocyte binding protein. Vaccine. 2000;19:508–13. doi: 10.1016/s0264-410x(00)00185-7. [DOI] [PubMed] [Google Scholar]
- 75.Lopez R, Urquiza M, Curtidor H, Eduardo Caminos J, Mora H, Puentes A, Patarroyo ME. Plasmodium falciparum: red blood cell binding studies of peptides derived from histidine-rich KAHRP-I, HRP-II and HRP-III proteins. Acta Trop. 2000;75:349–59. doi: 10.1016/s0001-706x(00)00071-1. [DOI] [PubMed] [Google Scholar]
- 76.Puentes A, Garcia J, Vera R, Lopez QR, Urquiza M, Vanegas M, Salazar LM, Patarroyo ME. Serine repeats antigen peptides which bind specifically to red blood cells. Parasitol Int. 2000;49:105–17. doi: 10.1016/s1383-5769(00)00040-4. [DOI] [PubMed] [Google Scholar]
- 77.Curtidor H, Rodriguez LE, Ocampo M, Lopez R, Garcia JE, Valbuena J, Vera R, Puentes A, Vanegas M, Patarroyo ME. Specific erythrocyte binding capacity and biological activity of Plasmodium falciparum erythrocyte binding ligand 1 (EBL-1)-derived peptides. Protein Sci. 2005;14:464–73. doi: 10.1110/ps.041084305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ocampo M, Vera R, Rodriguez LE, Curtidor H, Suarez J, Garcia J, Puentes A, Lopez R, Valbuena J, Tovar D, Reyes C, Vega S, Patarroyo ME. Identification of Plasmodium falciparum reticulocyte binding protein RBP-2 homologue a and b (PfRBP-2-Ha and -Hb) sequences that specifically bind to erythrocytes. Parasitol Int. 2004;53:77–88. doi: 10.1016/j.parint.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 79.Curtidor H, Ocampo M, Tovar D, Lopez R, Garcia J, Valbuena J, Vera R, Suarez J, Rodriguez LE, Puentes A, Guzman F, Torres E, Patarroyo ME. Specific erythrocyte binding capacity and biological activity of Plasmodium falciparum-derived rhoptry-associated protein 1 peptides. Vaccine. 2004;22:1054–62. doi: 10.1016/j.vaccine.2003.07.019. [DOI] [PubMed] [Google Scholar]
- 80.Curtidor H, Urquiza M, Suarez JE, Rodriguez LE, Ocampo M, Puentes A, Garcia JE, Vera R, Lopez R, Ramirez LE, Pinzon M, Patarroyo ME. Plasmodium falciparum acid basic repeat antigen (ABRA) peptides: erythrocyte binding and biological activity. Vaccine. 2001;19:4496–504. doi: 10.1016/s0264-410x(01)00202-x. [DOI] [PubMed] [Google Scholar]
- 81.Espejo F, Bermudez A, Torres E, Urquiza M, Rodriguez R, Lopez Y, Patarroyo ME. Shortening and modifying the 1513 MSP-1 peptide's alpha-helical region induces protection against malaria. Biochem Biophys Res Commun. 2004;315:418–27. doi: 10.1016/j.bbrc.2004.01.072. [DOI] [PubMed] [Google Scholar]
- 82.Espejo F, Cubillos M, Salazar LM, Guzman F, Urquiza M, Ocampo M, Silva Y, Rodriguez R, Lioy E, Patarroyo ME. Structure, immunogenicity, and protectivity relationship for the 1585 malarial peptide and its substitution analogues. Angew Chem Int Ed Engl. 2001;40:4654–7. doi: 10.1002/1521-3773(20011217)40:24<4654::aid-anie4654>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 83.Cubillos M, Salazar LM, Torres L, Patarroyo ME. Protection against experimental P falciparum malaria is associated with short AMA-1 peptide analogue alpha-helical structures. Biochimie. 2002;84:1181–8. doi: 10.1016/s0300-9084(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 84.Torres MH, Salazar LM, Vanegas M, Guzman F, Rodriguez R, Silva Y, Rosas J, Patarroyo ME. Modified merozoite surface protein-1 peptides with short alpha helical regions are associated with inducing protection against malaria. Eur J Biochem. 2003;270:3946–52. doi: 10.1046/j.1432-1033.2003.03780.x. [DOI] [PubMed] [Google Scholar]
- 85.Cifuentes G, Guzman F, Alba MP, Salazar LM, Patarroyo ME. Analysis of a Plasmodium falciparum EBA-175 peptide with high binding capacity to erythrocytes and their analogues using 1H NMR. J Struct Biol. 2003;141:115–21. doi: 10.1016/s1047-8477(02)00584-1. [DOI] [PubMed] [Google Scholar]
- 86.Cifuentes G, Patarroyo ME, Urquiza M, Ramirez LE, Reyes C, Rodriguez R. Distorting malaria peptide backbone structure to enable fitting into MHC class II molecules renders modified peptides immunogenic and protective. J Med Chem. 2003;46:2250–3. doi: 10.1021/jm020440w. [DOI] [PubMed] [Google Scholar]
- 87.Purmova J, Salazar LM, Espejo F, Torres MH, Cubillos M, Torres E, Lopez Y, Rodriguez R, Patarroyo ME. NMR structure of Plasmodium falciparum malaria peptide correlates with protective immunity. Biochim Biophys Acta. 2002;1571:27–33. doi: 10.1016/s0304-4165(02)00203-9. [DOI] [PubMed] [Google Scholar]
- 88.Salazar LM, Alba MP, Torres MH, Pinto M, Cortes X, Torres L, Patarroyo ME. Protection against experimental malaria associated with AMA-1 peptide analogue structures. FEBS Lett. 2002;527:95–100. doi: 10.1016/s0014-5793(02)03174-5. [DOI] [PubMed] [Google Scholar]
- 89.Alba MP, Salazar LM, Vargas LE, Trujillo M, López Y, Patarroyo ME. Modifying RESA protein peptide 6671 to fit into HLA-DRbeta1* pockets induces protection against malaria. Biochem Biophys Res Commun. 2004;315:1154–64. doi: 10.1016/j.bbrc.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 90.Alba MP, Salazar LM, Puentes A, Pinto M, Torres E, Patarroyo ME. 6746 SERA peptide analogues immunogenicity and protective efficacy against malaria is associated with short alpha helix formation: malaria protection associated with peptides alpha helix shortening. Peptides. 2003;24:999–1006. doi: 10.1016/s0196-9781(03)00187-6. [DOI] [PubMed] [Google Scholar]
- 91.Patarroyo ME, Cifuentes G, Vargas LE, Rosas J. Structural modifications enable conserved peptides to fit into MHC mole-cules thus inducing protection against malaria. Chembiochem. 2004;5:1588–93. doi: 10.1002/cbic.200400116. [DOI] [PubMed] [Google Scholar]
- 92.Patarroyo ME, Alba MP, Vargas LE, Silva Y, Rosas J, Rodriguez R. Peptides inducing short-lived antibody responses against Plasmodium falciparum malaria have shorter structures and are read in a different MHC II functional register. Biochemistry. 2005;44:6745–54. doi: 10.1021/bi050214z. [DOI] [PubMed] [Google Scholar]
- 93.Patarroyo ME, Bermudez A, Salazar LM, Espejo F. High non-protective, long-lasting antibody levels in malaria are associated with haplotype shifting in MHC-peptide-TCR complex formation: a new mechanism for immune evasion. Biochimie. 2006;88:775–84. doi: 10.1016/j.biochi.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 94.Rammensee HG, Friede T, Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 95.Reyes C, Patarroyo ME, Vargas LE, Rodriguez LE, Patarroyo MA. Functional, structural, and immunological compartmentalisation of malaria invasive proteins. Biochem Biophys Res Commun. 2007;354:363–71. doi: 10.1016/j.bbrc.2006.12.220. [DOI] [PubMed] [Google Scholar]
- 96.Blackman MJ, Holder AA. Secondary processing of the > merozoite surface protein-1 (MSP1) by a calcium-dependent membrane-bound serine protease: shedding of MSP133 as a noncovalently associated complex with other fragments of the MSP1. Mol Biochem Parasitol. 1992;50:307–15. doi: 10.1016/0166-6851(92)90228-c. [DOI] [PubMed] [Google Scholar]
- 97.Pachebat JA, Kadekoppala M, Grainger M, Dluzewski AR, Gunaratne RS, Scott-Finnigan TJ, Ogun SA, Ling IT, Bannister LH, Taylor HM, Mitchell GH, Holder AA. Extensive proteolytic processing of the malaria parasite merozoite surface protein 7 during biosynthesis and parasite release from erythrocytes. Mol Biochem Parasitol. 2007;151:59–69. doi: 10.1016/j.molbiopara.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 98.Li J, Mitamura T, Fox BA, Bzik DJ, Horii T. Differential localization of processed fragments of Plasmodium falciparum serine repeat antigen and further processing of its N-terminal 47 kDa fragment. Parasitol Int. 2002;51:343–52. doi: 10.1016/s1383-5769(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 99.Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, Lopez-Estrano C, Haldar K. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–7. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 100.Andersen CA, Palmer AG, Brunak S, Rost B. Continuum secondary structure captures protein flexibility. Structure. 2002;10:175–84. doi: 10.1016/s0969-2126(02)00700-1. [DOI] [PubMed] [Google Scholar]
- 101.Hennecke J, Wiley DC. Structure of a complex of the human alpha/beta T cell receptor (TCR) HA1.7, influenza hemag-glutinin peptide, and major histocompati-bility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): insight into TCR cross-restriction and alloreactivity. J Exp Med. 2002;195:571–81. doi: 10.1084/jem.20011194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Svensson AC, Setterblad N, Pihlgren U, Rask L, Andersson G. Evolutionary relationship between human major histocompatibility complex HLA-DR haplotypes. Immunogenetics. 1996;43:304–14. doi: 10.1007/BF02440998. [DOI] [PubMed] [Google Scholar]
- 103.Dessen A, Lawrence CM, Cupo S, Zaller DM, Wiley DC. X-ray crystal structure of HLA-DR4 (DRA*0101, DRB1*0401) complexed with a peptide from human collagen II. Immunity. 1997;7:473–81. doi: 10.1016/s1074-7613(00)80369-6. [DOI] [PubMed] [Google Scholar]
- 104.Hennecke J, Carfi A, Wiley DC. Structure of a covalently stabilized complex of a human alphabeta T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. EMBO J. 2000;19:5611–24. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reinherz EL, Tan K, Tang L, Kern P, Liu J, Xiong Y, Hussey RE, Smolyar A, Hare B, Zhang R, Joachimiak A, Chang HC, Wagner G, Wang J. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 1999;286:1913–21. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 106.Jones EY, Fugger L, Strominger JL, Siebold C. MHC class II proteins and disease: a structural perspective. Nat Rev Immunol. 2006;6:271–82. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 107.Cardenas C, Villaveces JL, Bohorquez H, Llanos E, Suarez C, Obregon M, Patarroyo ME. Quantum chemical analysis explains hemagglutinin peptide-MHC Class II molecule HLA-DRbeta1*0101 interactions. Biochem Biophys Res Commun. 2004;323:1265–77. doi: 10.1016/j.bbrc.2004.08.225. [DOI] [PubMed] [Google Scholar]
- 108.Cardenas C, Villaveces JL, Suarez C, Obregon M, Ortiz M, Patarroyo ME. A comparative study of MHC Class-II HLA-DRbeta1*0401-Col II and HLA-DRbeta1*0101-HA complexes: a theoretical point of view. J Struct Biol. 2005;149:38–52. doi: 10.1016/j.jsb.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 109.Bermudez A, Cifuentes G, Guzman F, Salazar LM, Patarroyo ME. Immunogeni-city and protectivity of Plasmodium falciparum EBA-175 peptide and its analog is associated with alpha-helical region shortening and displacement. Biol Chem. 2003;384:1443–50. doi: 10.1515/BC.2003.160. [DOI] [PubMed] [Google Scholar]


