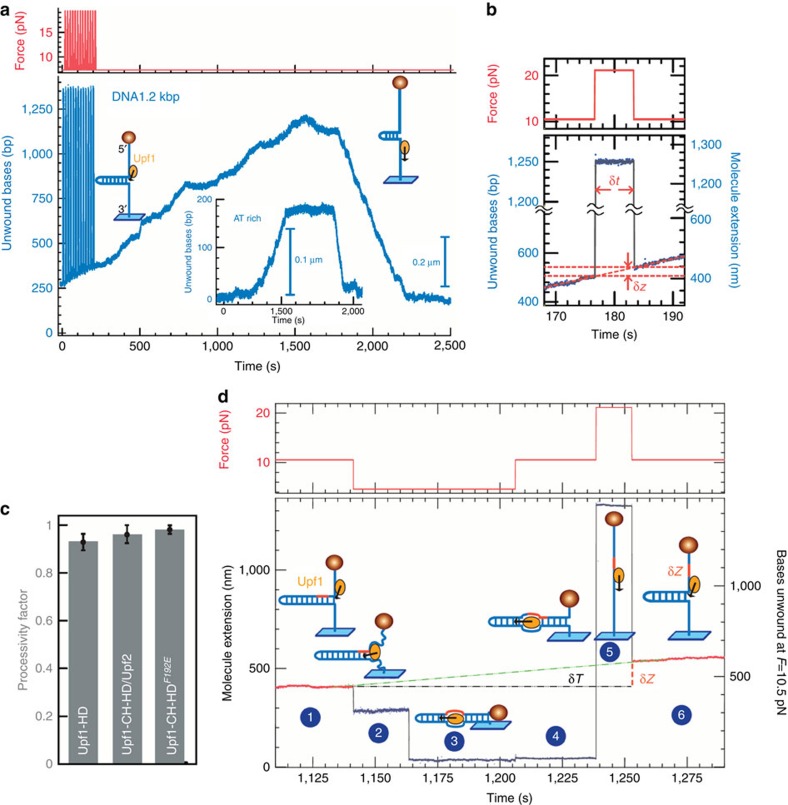
Figure 2. Single-molecule characterization of Upf1 processivity.
(a) Trace obtained with a 1.2 kb DNA hairpin. A series of force surges (from 0 to 250 s) were applied to the hairpin, exposing single strand, favouring Upf1 binding. This saw-tooth event consists of unwinding (250 and 1,600 s) and ssDNA translocation (1,600 and 2,230 s) of Upf1-HD. (Inset) Similar event with a 180-bps AT-rich DNA hairpin. The GC-rich sequence close to the apex leads to pauses (1,530 and 1,800 s). (b) Single strand translocation process observed during a force-jump assay. The amount δz that Upf1-HD has travelled during the force surge lasting time interval δt is read when the hairpin refold. (c) Relative processivity of fully active Upf1 forms represented by the processivity factor: Upf1-HD fp=52/56, Upf1-CH-HD/Upf2 fp=24/25, Upf1F192E fp=55/56, error bars are s.d. (d) Force modulation (red) in time and molecule extension (blue) showing that Upf1 is active in a DNA bubble. In phases 1 and 5 Upf1-HD unwinds at 10 pN. In phases 2 and 3, F was reduced to 5 pN (1,140 and 1,240 s). During a transient (1,140 and 1,164 s) the helicase still blocked the fork, and in phase 3 the unwound ssDNA arms passed over the helicase so that the hairpin fully refolds, enclosing Upf1-HD within a bubble. In phase 4, a force surge to 21 pN (1,240 and 1,252 s) fully re-opens the hairpin. In phase 5, reducing F to 10 pN refolds the hairpin partially since the fork is blocked again by Upf1-HD. During the entire time interval (δT) the helicase had slowly progressed within the DNA bubble and was not expelled.