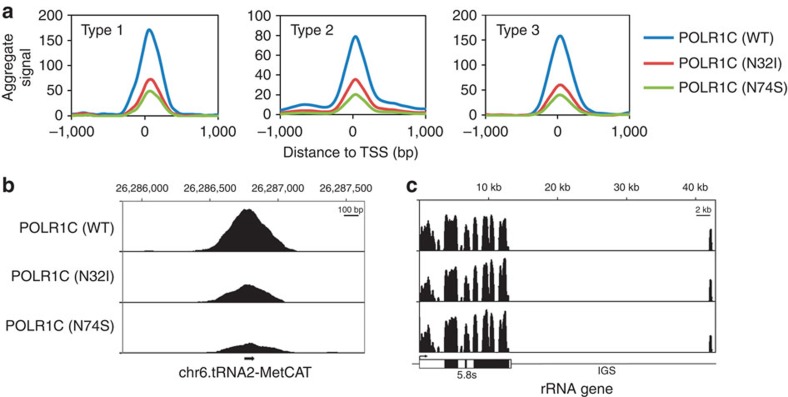
Figure 4. Impact of POLR1C mutations on polymerase association with chromatin.
(a) ChIP-Seq experiments of FLAG-tagged POLR1C variants (wild type, N32I or N74S). Aggregate profile produced with the annotation mode of the Versatile Aggregate Profiler shows ChIP-Seq data sets over the three classes of POLR3-transcribed genes, as defined by the promoter structure. Type 1 genes have an internal promoter composed of A and C boxes (5S rRNA genes). Type 2 genes have an internal promoter composed of A and B boxes (tRNA genes, for example), while the promoter of type 3 genes (U6, 7SK, RNase P and others) is located upstream of the transcription start site (TSS)55. The TSS was used as the reference point. (b) IGV view of a tRNA-Met gene transcribed by POLR3. POLR1C binding is decreased in mutated variants compared with wild type. IGS, intergenic spacer. (c) IGV view of one ribosomal DNA (rDNA) repeat. The rDNA gene encodes a 45S pre-rRNA precursor that will generate the 5.8S, 18S and 28S rRNAs. There are ∼400 copies of the rDNA gene arranged in tandem repeats in the human genome. rDNA repeats are not present in the reference genome assemblies; therefore, unique reads were aligned directly to the human rDNA reference sequence (NCBI accession number: HSU13369)56. No differences were observed in POLR1C occupancy between wild-type and mutant variants. A schematic of a rDNA repeat is included below the graph.