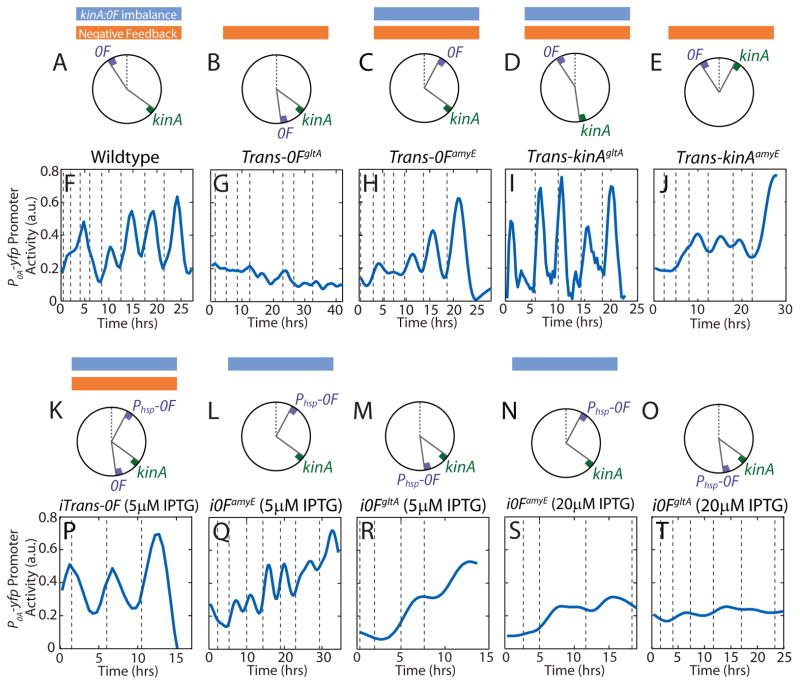
Figure 4.
Experimental observation confirm that 0A~P pulsing depends on kinA-0F chromosomal arrangement and transcriptional feedback from 0A~P to 0F negative feedback. A–E Chromosomal arrangements of 0F and kinA in Wildtype B. subtilis (A), Trans-0FgltA (B), Trans-0FamyE (C), Trans-kinAgltA (D) and Trans-kinAamyE (E) strains. Blue and orange bars mark strains that have transient kinA:0F imbalance and negative feedback respectively. F–J Measurements of P0A-yfp activity in single cells during starvation. 0A~P pulses seen in Wildtype B. subtilis cells (F) are abolished by the translocation of 0F to the gltA locus in the Trans-0FgltA strain (G) but not by the translocation of 0F to the amyE locus in the Trans-0FamyE strain (H). 0A~P pulses are also abolished by the translocation of kinA to the amyE locus in the Trans-kinAgltA strain (J) but not by the translocation of kinA to the gltA locus in the Trans-0FgltA strain (I). Note that P0A-yfp promoter activity level does not increase in the Trans-0FgltA strain. Vertical dashed lines indicate cell divisions. K–O Chromosomal arrangements of 0F and kinA in iTrans-0F (K), i0FamyE (L, N) and i0FgltA strains (M, O). Note that both i0F strains lack the 0A~P-0F negative feedback. P–T Measurements of P0A-yfp activity in single cells during starvation. Addition of an IPTG inducible copy of 0F near the origin in the iTrans-0F strain (P) recovers the transient kinA:0F imbalance lost in Trans-0FgltA strain and rescues the 0A~P pulses seen in wildtype B. subtilis cells. 0A activity pulsing is greatly decreased in the i0FamyE (Q, S) and i0FgltA strains (R, T) which lack the negative feedback. 0A activity in the i0FamyE (Q) strain fluctuates due to transient changes in kinA:Phsp-0F but does not pulse (Figure S5). If 0F expression is low (at 5μM IPTG; Q, R), both i0F strains accumulate high 0A activity levels. High level expression of 0F (at 20μM IPTG; S, T) blocks 0A activation in both i0F strains similar to Trans-0FgltA.