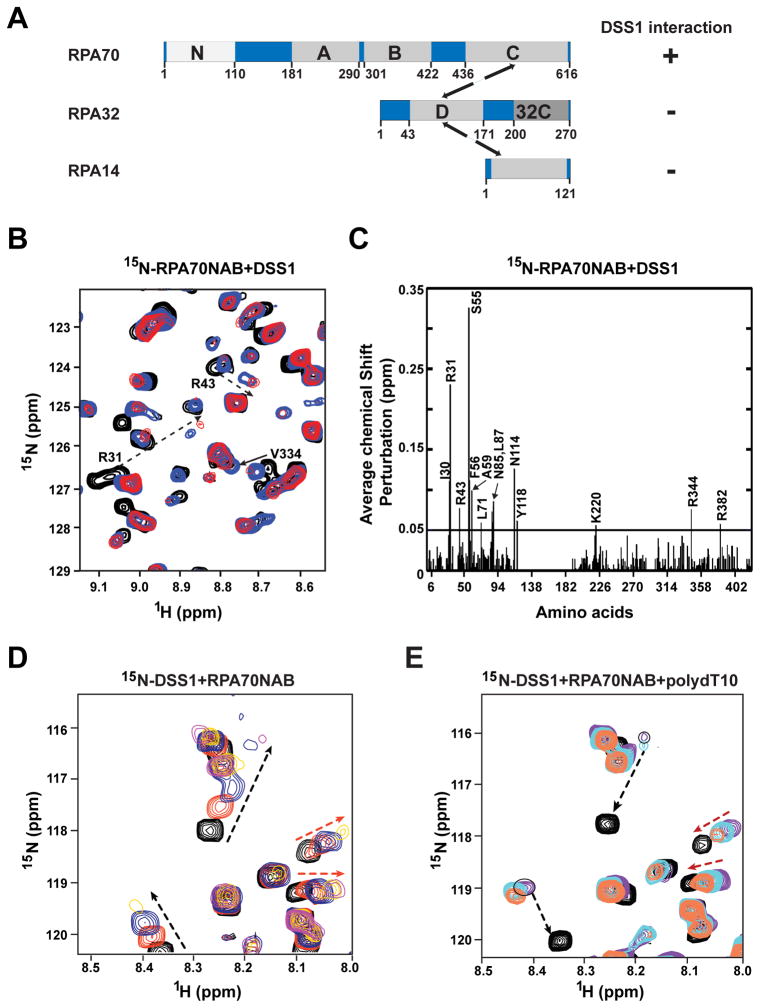
Figure 3. Evidence for RPA70 interaction activity and DNA mimicry in DSS1.
See also Figure S2 and Figure S3.
(A) Schematic of the RPA subunits RPA70, RPA32, and RPA14 with functional domains.
(B) Overlay of 15N-1HSQC spectra of 15N-enriched RPA70NAB as the solution was titrated with DSS1. The circles identify peaks belonging to RPA70N. The solid and dotted arrows indicate significant chemical shift perturbations upon binding of DSS1 for residues within RPA70AB and 70N, respectively.
(C) Chemical shift changes plotted against the sequence of RPA70NAB. The mean value of chemical shift changes is shown by the solid horizontal line.
(D) Overlay of 15N-1HSQC spectra of 15N-enriched DSS1 in complex with unlabeled RPA70NAB at molar ratios of 1:0 (black), 1:0.2 (red), 1:0.4 (blue), and 1:1 (yellow). The black and red dotted arrows identify peaks with chemical shift preturbations arsing from interaction with 70N and with 70AB, respectively.
(E) Overlay of 15N-1HSQC spectra of 15N-enriched DSS1 in complex with unlabeled RPA70NAB and ssDNA. Spectra were acquired at DSS1:RPA70NAB:ssDNA ratios of 1:0:0 (black), 1:1:0 (purple), 1:1:0.5 (cyan) and 1:1:1 (red) molar ratios. Red arrows identify two peaks that revert to the position of free DSS1 upon addition of ssDNA due to release of DSS1 by ssDNA binding to RPA70AB. Black arrows identify two peaks that are not affected by addition of ssDNA because they interact with RPA70N.