Table 1. Kinetic and thermodynamic parameters for CueR−DNA interactions in E. coli cells*.
| Parameters | 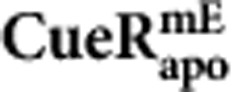 |
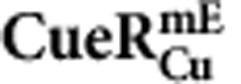 |
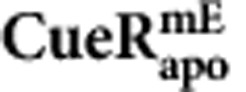 (dividing cells)
(dividing cells)
|
|---|---|---|---|
| k1 (μM−1 s−1) | 214±46 | 54±96 | 219±77 |
| k−1 (s−1) | 8.2±0.9 | 4.6±0.8 | 6.4±1.1 |
| kf (μM−1 s−1) | 31.9±6.9 | 55.0±8.5 | 20.1±8.7 |
| KD1(k−1/k1) (μM) | 0.037±0.028 | 0.038±0.058 | 0.029±0.011 |
| k2 (μM−1 s−1) | 3.6±1.9 | 4.9±3.8 | 1.6±6.1 |
| k−2 (s−1) | 2.5±0.1 | 4.1±0.1 | 2.6±0.2 |
| KD2(=k−2/k2) (μM) | 0.69±0.38 | 0.83±0.65 | 1.6±6.3 |
| KD3(=k−3/k3) | 0.06±0.03 | 0.04±0.05 | 0.02±0.08 |
| NNB | 2,605±1,381 | 2,588±1,976 | 8,941±34,406 |
| NSB | 130±17 | 121±89 | 153±36 |
*See Fig. 2e for definition of kinetic parameters. NNB and NSB are the effective numbers of specific recognition sites and nonspecific binding sites on the chromosome. All error bars are s.d. Relevant data for ZntR variants and other control strains are in Supplementary Tables 7 and 8.
