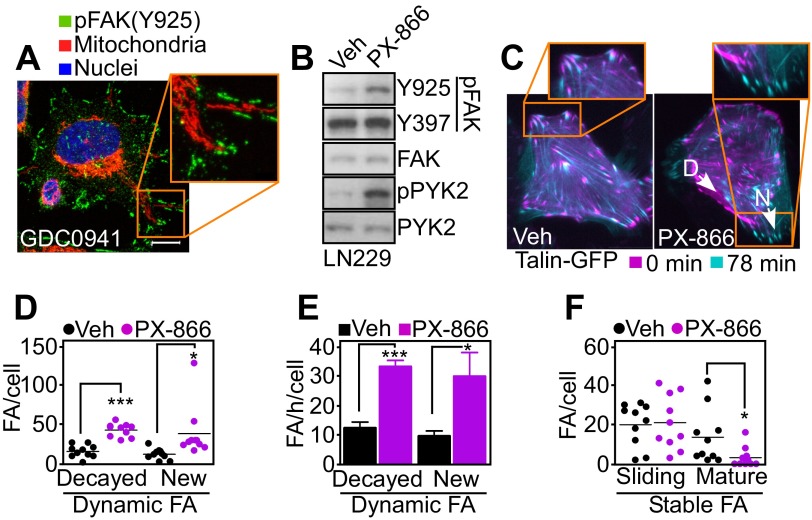
Fig. S5.
PI3K therapy regulation of focal adhesion dynamics. (A) PC3 cells treated with the PI3K inhibitor GDC0941 for 48 h were replated onto fibronectin-coated slides for 5 h and labeled with an antibody to phosphorylated FAK (pY925) Alexa488, MitoTracker Red, or DAPI. Representative 1-µm extended-focus confocal images with localization of mitochondria near pFAK+ focal adhesion complexes are shown. Magnification, 63×. Inset was zoomed digitally at 10×. (Scale bar, 10 μm.) (B) LN229 cells were treated with vehicle or PX-866 for 48 h and protein extracts were analyzed by Western blotting. p, phosphorylated. (C) LN229 cells expressing Talin-GFP to label FA complexes were treated with vehicle or PX-866 for 48 h and analyzed by time-lapse microscopy. Representative images at initial (0 min) and final time points (78 min) were merged to visualize FA turnover. Arrows indicate representative decayed (D) or new (N) FAs. Magnification, 40×. (D–F) Individual FA complexes in LN229 cells expressing Talin-GFP as in C were manually tracked to determine FA dynamics. (D) Number of dynamic FA complexes per cell classified into disappearing (decayed) and newly formed (new). Mean ± SEM (n = 10). *P = 0.0353; ***P < 0.0001. (E) Turnover rates (number of FA complexes per h) of FA decay and formation. Mean ± SEM (n = 10). *P = 0.0235; ***P < 0.0001. (F) Stable FA complexes classified as sliding (moving) or mature (>50% overlay between initial and final localization). Mean ± SEM (n = 10). *P = 0.0393.