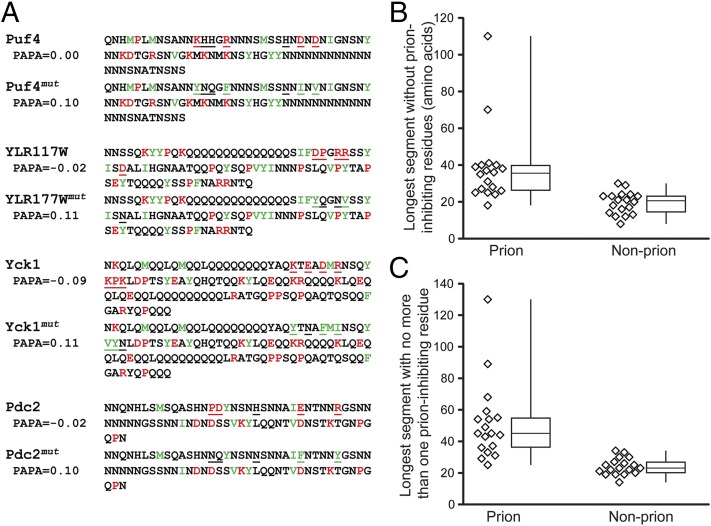
Fig. 1.
Design of prion-promoting mutations. (A) Sequences of the wild-type and mutant Puf4, YLR177W, Yck1, and Pdc2 PrLDs. Strongly prion-promoting amino acids (W, Y, F, V, I, and M) are indicated in green, whereas strongly prion-inhibiting amino acids (P, K, R, D, and E) are in red. Positions that were mutated are underlined. (B) PrLDs that do not show detectable prion activity tend to lack extended peptide stretches without prion-inhibiting residues. Alberti et al. (14) identified 100 yeast fragments with prion-like composition and tested each in four assays for prion-like activity. Shown are box-and-whiskers plots of the longest stretch without any prion-inhibiting residues for each of the proteins that showed prion-like activity in all four assays (Prion) and each of the proteins that failed all four assays (Nonprion). (C) Box-and-whiskers plot of the longest segments with no more than one prion-inhibiting residue.