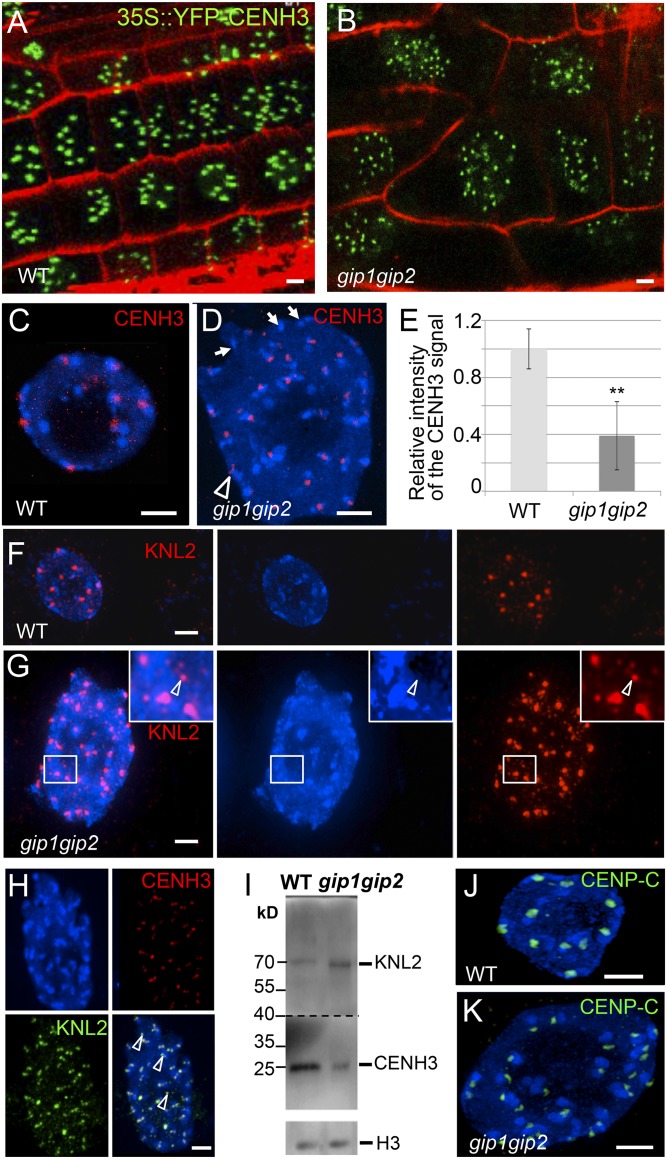
Fig. 4.
gip1gip2 mutants exhibit defects of centromeric components in cycling cells. 35S::YFP-CENH3 expression in root tips from WT (A) and gip1gip2 (B) using identical imaging settings. (C–H) Immunolocalization of CENH3 and/or KNL2 in WT (C and F) and gip nuclei (D, G, and H). DAPI staining is shown in blue. (D) Arrowhead(s) and arrows indicate elongated signal(s) or absence of signal, respectively. (E) Mean intensity of the CENH3 signals: WT, n = 50; gip mutants, n = 156. **P < 0.01. (G) The arrowhead shows a KNL2 signal outside of a chromocenter. (H) Arrowheads in the merged image indicate KNL2 signals not colocalized with CENH3 in the nucleoplasm. (I) Western blot analysis performed on the same blot of endogenous CENH3 and KNL2 protein amounts in nuclear extracts from WT and gip1gip2. Histone H3 was used as a loading control. This experiment was reproduced four times. (J and K) Immunolocalization of CENP-C in WT (J) and gip nuclei (K). (Scale bars, 2 µm.)