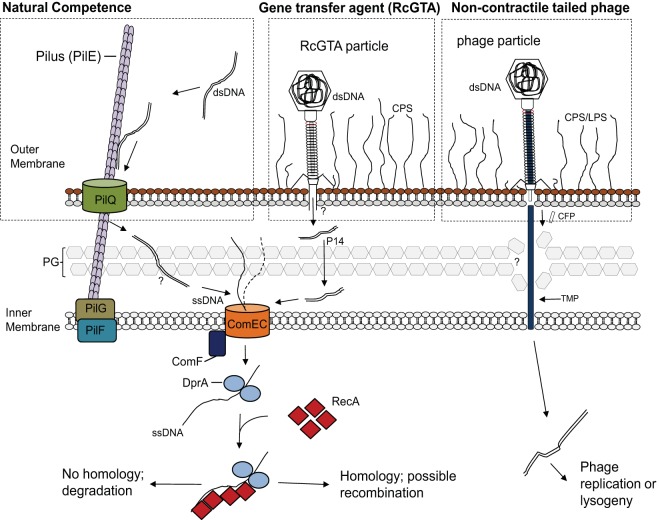
FIG 5.
Schematic diagram comparing an overview of natural transformation systems in Gram-negative bacteria to the proposed RcGTA recipient capability pathway and a noncontractile tailed phage pathway. The conserved steps in natural transformation are shown on the left. DNA is taken up by a transformation-dedicated pilus (Tfb), which binds dsDNA and brings it into the periplasm, where it is processed into ssDNA by an unknown process. DNA then enters the cytoplasm via the ComEC inner membrane protein in a pathway involving ComF. Upon cytoplasmic internalization, ssDNA is bound by DprA, which functions to recruit the RecA recombinase to polymerize onto ssDNA, promoting a homology search and subsequent recombination into the chromosome (14). The proposed model of RcGTA, shown in the center, involves the same steps for DNA import and recombination into the chromosome as those in natural transformation. It is unknown how RcGTA-injected DNA traverses the periplasm or whether DNA enters the cell as ssDNA or dsDNA. Passage through the peptidoglycan (PG) likely is facilitated by the RcGTA protein p14, which degrades PG (46). Shown on the right is a model of noncontractile tailed phage infection, with the tape measure protein (TMP) used to traverse the outer membrane, periplasm, and cytoplasmic membrane (50). CFP, central tail fiber protein; LPS, lipopolysaccharide; CPS, capsular polysaccharide. (The natural transformation pathway image was adapted from reference 14 with permission of the publisher.)