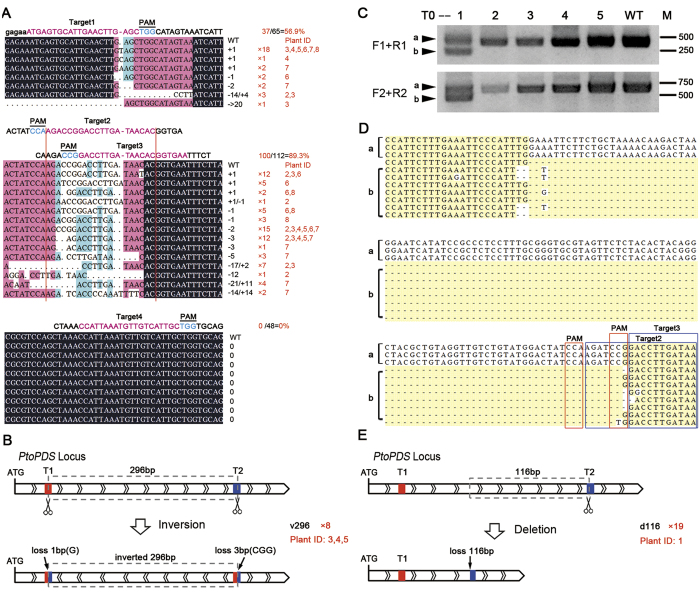
Figure 3. CRISPR/Cas9-mediated gene editing in transgenic poplar plants.
(A) A range of insertion and deletions (indels) were found in the other T0 plants. Red number in right side means the no. of detected clones with such mutant allele and in which plant such mutant allele is found. (B) A big fragment inversion mutant of PtoPDS. The sequence between the target site1 and target site 2 (296 bp) of PtoPDS were cut down by Cas9 and ligated back to the genome reversely, with 1 and 3 bp of nucleotides around the up- and downstream cut sites were lost. (C) DNA samples from independent transgenic poplar were analyzed for mutations by PCR assays. In the top row, F1 and R1 were used; in the bottom row, F2 and R2 were used. T0-1 represents a heterozygous mutant of PDS, in which two amplification bands with different length were obtained in this assay: “a” indicates the full-length fragment of the PDS gene, “b” indicates a shorter fragment of the PDS gene with mutation. (D) The PCR products from T0-1 plant were separated by their size and cloned and sequenced to validate the deletion directed by CRISPR/Cas9 in the PDS gene. (E) A big fragment deletion mutant of PtoPDS. The 116 bp sequence upstream of target site 2 was lost in the DNA repairing after Cas9 mediated double strain break. In B and E, Red and blue quadrangles indicate the target site 1 and 2 in PtoPDS locus respectively; the red number in the right side means the no. of detected clones with such mutant allele.