Fig. 1.
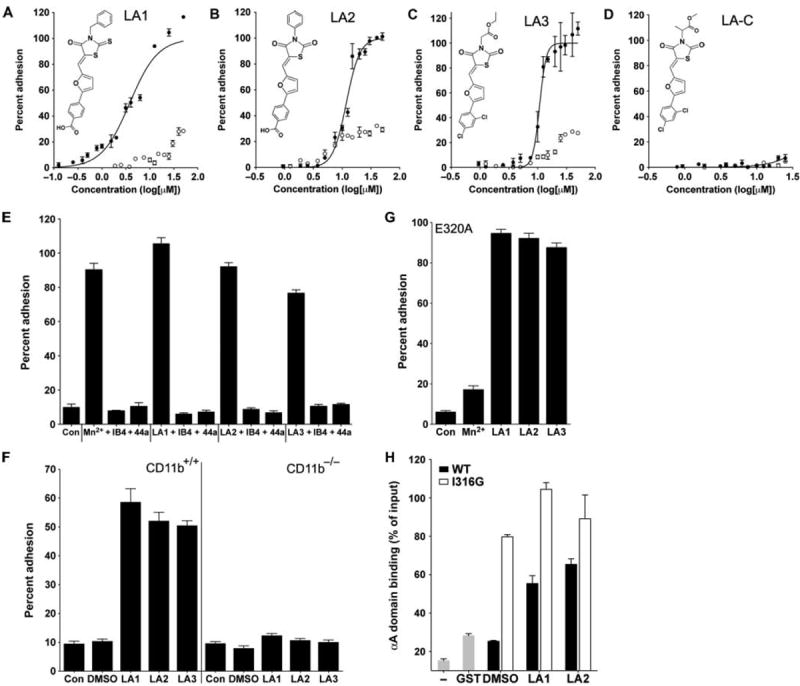
Leukadherins increase CD11b/CD18-dependent cell adhesion. (A to D) Dose-response curves showing the percentages of input K562 CD11b/CD18 cells (filled circles) and K562 cells (open circles) that adhered to immobilized fibrinogen in the presence of increasing amounts of LA1, LA2, LA3, or LA-C. The chemical structures of LA1, LA2, LA3, and LA-C are also shown. Data are means ± SEM (n = 3 to 6 replicates per data point) from one of at least three independent experiments. (E) Histograms showing the adhesion of K562 CD11b/CD18 cells to fibrinogen in response to LA1, LA2, or LA3 in the absence or presence of the blocking antibodies IB4 and 44a. Also shown is the extent of adhesion in the presence of physiologic concentrations of Ca2+ and Mg2+ (Con) and that in the presence of the known agonist Mn2+. Data are means ± SEM (n = 4 to 9 replicates per condition) from one of at least three independent experiments. (F) Histograms showing the percentage adhesion of wild-type (WT, CD11b+/+) and CD11b−/− neutrophils to immobilized fibrinogen in the absence (DMSO) or presence of LA1, LA2, or LA3 as compared to that of control cells (Con). Data are means ± SEM (n=5 replicates per condition) from one of at least three independent experiments. (G) Histograms showing the percentage of binding of K562 E320A cells to immobilized fibrinogen induced by LA1, LA2, or LA3. Also shown is the extent of K562 E320A adhesion with Ca2+ and Mg2+ ions (Con) and Mn2+. Data are means ± SEM (n = 6 replicates per condition) from one of at least three independent experiments. (H) Histograms showing the percentage binding of recombinant GST-fused αA domain constructs to immobilized fibrinogen (normalized to the amounts of input αA) in the absence of leukadherin (DMSO) or in the presence of LA1 or LA2. Also shown is the background binding obtained in the absence of any protein (−) or with the GST construct alone (GST). Data are means ± SEM (n = 3 replicates per condition) from one of at least three independent experiments.
