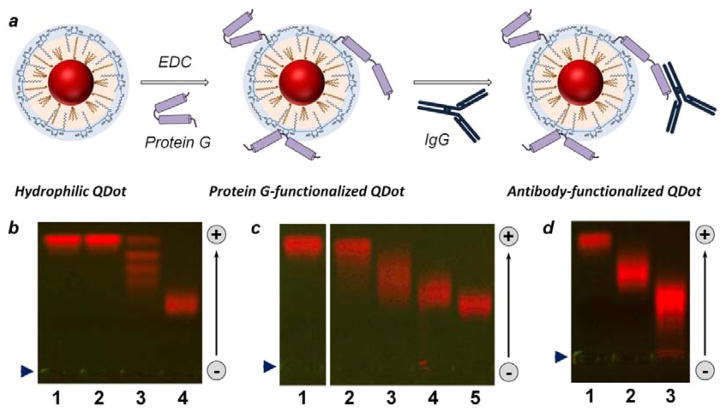
Figure 4.
Bioconjugation of polymer-encapsulated QDots. (a) Schematic of QDot-PrG covalent bioconjugation and QDot-PrG-IgG non-covalent self-assembly procedure for preparation of targeted QDot probes. (b–d) Characterization of quantum dot biofunctionalization with agarose gel electrophoresis. (b) Covalent conjugation of highly negatively-charged QDots with uncharged SA (lane 3) and PrG (lane 4) resulted in reduction in QDot gel motility. Interestingly, agarose gel produced distinct “ladder” for QDots with different number of SA molecules attached in lane 3, but failed to resolve bioconjugation stoichiometry for QDot-PrG conjugates, which migrated as a single broader band in lane 4. Reference unconjugated QDots were loaded to lane 1, and control QDots treated with EDC alone were loaded to lane 2. (c) Bioconjugation with increasing PrG:QDot excess from 1:1 (lane 2) to 5:1 (lane 3), 10:1 (lane 4), and 20:1 (lane 5) deposited increasing number of PrG molecules onto each QDot, as evident from slow-down in gel motility of QDot-PrG bioconjugates. Reference unconjugated QDots were loaded to lane 1. (d) QDot functionalization with an IgG via PrG-IgG binding further reduced QDot-PrG-IgG probe motility (lane 3) in comparison to QDot-PrG probes (lane 2) and unconjugated QDots (lane 1). Loading well position is indicated by blue arrowhead on the left. Migration of QDots in the gel is indicated by an arrow on the right.