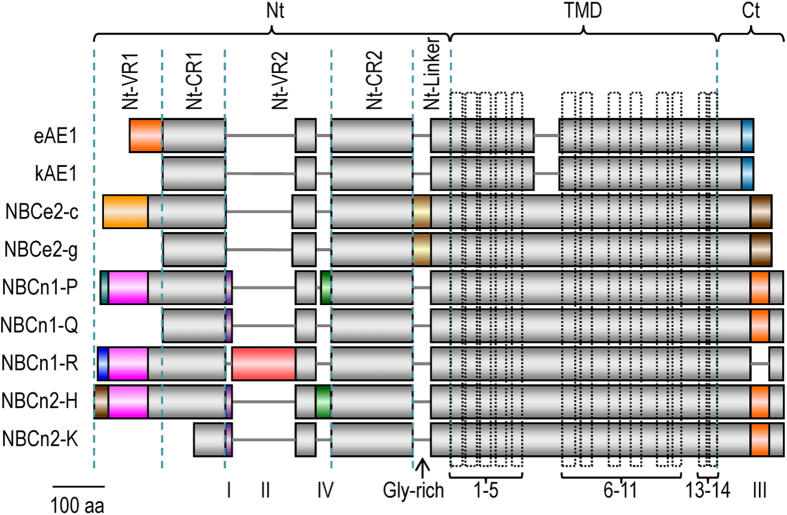
Figure 14. Structural comparison of variants of selected Slc4 members.
The diagram was drawn based upon a sequence alignment for rat eAE1 (accession #NP_036783.2), rat kAE1 (accession #AAH85748.1), rat NBCe2-c (accession #NP_997677.1), rat NBCe2-g (accession #BAM73282.1), mouse NBCn1-P (accession #AFI43932.1), mouse NBCn1-Q (accession #AGX13876.1), mouse NBCn1-R (accession #AGX13877.1), rat NBCn2-H (accession #JX073718), rat NBCn2-K (accession #AHG54969.1). The sequence alignment was performed with online multiple sequence alignment tool Clustal Omega from European Bioinformatics Institute of EMBL (http://www.ebi.ac.uk/Tools/msa/clustalo/). The Nt domains of the Slc4 transporters consist of two variable regions VR1 and VR2, as well as two conserved regions CR1 and CR2. Cassettes I through IV indicate the four optional structural elements of NBCn1. Compared to eAE1, kAE1 is truncated by 78 aa at the Nt. Compared to NBCe2-c, NBCe2-g is truncated by 115 aa. NBCn1-Q is truncated by 125 aa compared to NBCn1-P. NBCn2-K is truncated by 194 aa compared to NBCn2-H.