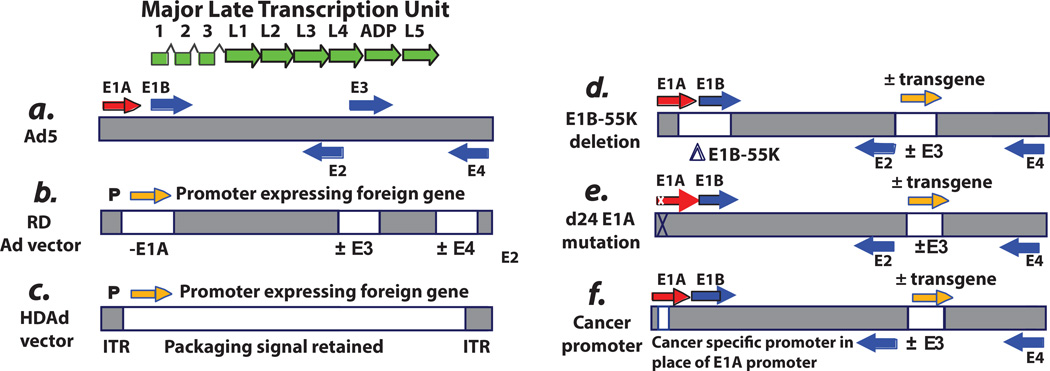
Fig. 1. Schematic of the genome of adenovirus type 5 (Ad5) and Ad5-based vectors.
A: Ad5. The horizontal bar indicates the duplex DNA genome of 36 kbp encoding ca. 35 genes. The arrows indicate transcription units. The “immediate early” E1A proteins derived from the E1A transcription unit (red arrow) induce expression of the “delayed early” proteins coded by the E1B, E2, E3, and E4 transcription units (blue arrows). Viral DNA begins to replicate at about 7 hours postinfection (p.i.), and then “late” proteins derived from the major late transcription unit (green arrows) are synthesized. The major late messenger RNAs (mRNAs) are formed by alternative splicing and polyadenylation of a large pre-mRNA initiated at the single major late promoter and extending to the right end of the genome. All late mRNAs have a tripartite leader (leaders 1, 2, and 3) at their 5′ termini that facilitates translation. Beginning at 20 to 24 hours p.i., virions begin to assemble in the cell nucleus, and then after 2 to 3 days the cells begin to lyse and release virions, with lysis complete by about 5 to 6 days. Efficient cell lysis is mediated by Adenovirus Death Protein (ADP). ADP is a late protein derived from the major late transcription unit. The vectors depicted are based on the Ad5 backbone. However, vectors based on serotypes other than Ad5 have also been developed. The reason for this is that Ad5 uses the Coxsackie-Adenovirus Receptor (CAR) receptor, whereas some other serotypes use different receptors (e.g., CD46 and sialic acid as receptors), and sometimes this is an advantage. Also, pre-existing antibody levels in humans are more prevalent for Ad5 than for certain other serotypes. B: Replication-defective vectors. The E1A and E1B regions (transcription units) (i.e., the E1 region) are deleted and replaced with an expression cassette with an exogenous promoter driving expression of the therapeutic gene. In gene therapy vectors, this gene can be one to correct a genetic defect. In vaccine vectors, the gene is the antigen. In cancer gene therapy vectors, the gene (e.g., p53) induces cell death. Usually the E3 region is deleted. Deletion of E3 does not affect the growth of the vector because the E3 genes are not required for Ad growth in cell culture. Also, deletion of E3 allows for larger inserts into the E1 region because only up to 105% of the genome can be packaged into virions. E1-deleted vectors are defective for replication because the E1A proteins, and in some cells the E1B proteins, are required for virus growth. The vectors are propagated in cell lines such as 293 or PER.C6, which retain and express the E1A and E1B proteins. Although the E1A proteins are required for expression of other Ad genes, these vectors can be leaky and express other Ad genes in an E1A-independent manner, especially at long periods following infection and if high multiplicities of infection are used. In animal model studies, this leakiness has led to elimination of vector-transduced cells by cells of the immune system. For this reason, second-generation vectors also lack the E4 region. E4 regions are essential for Ad replication, including late gene expression, so deletions of E4 eliminate the leakiness. Some vectors also lack the E2 region. These E4- and E2-deleted vectors must be grown on cell lines that complement the E1, E4, and E2 deletions. C: Helper-dependent vectors. These vectors lack all or most Ad genes but retain the cis-acting sequences such as the inverted terminal repeats (ITRs) at each end of the genome as well as the packaging sequence at the left end that are required for the genome to replicate and be packaged. These vectors are propagated in the presence of a helper Ad, which must be eliminated from the large-scale vector stocks. It is difficult to completely eliminate the vector. D: ONYX-015 and related vectors. ONYX-015 was the first replication-competent (oncolytic) Ad vector to be described. It lacks the E1B-55K gene as well as the rid and 14.7k genes in the E3B region. (ONYX-015 does not have a transgene in E3). Because of the E1B-55K deletion, this vector grows better in cancer cells than in noncancerous cells. However, in many cancer cells, the vector does not grow as well as Ad5. Many other oncolytic Ad vectors have a similar design (e.g., Oncorine), and some vectors have a therapeutic gene (orange arrow) (e.g. GM-CSF) incorporated into the E3 region and expressed from the E3 promoter, the major late promoter via splicing, or from a promoter inserted into E3. E: D24 vectors with an E1A CR2 deletion. Some vectors are targeted to cancer cells by virtue of deletions in the el a gene that knock out binding of the E1A proteins to the pRB family of tumor suppressors. Vectors with the CR2 deletion are able to replicate well in cancer cells with a defect in the pRB tumor suppressor pathway. The vectors replicate less well in normal cells. F: Vectors with a cancer-specific promoter replacing the E1A enhancer/promoter. Many vectors are targeted to cancer cells by replacement of the E1A promoter/enhancer by a cancer-specific promoter. These vectors grow in cancer cells in which the promoter is active. Many different promoters have been used. Some vectors have a therapeutic transgene in E3 and some vectors have an intact E3 region. In some vectors, the E4 promoter is replaced by a cancer-specific promoter.