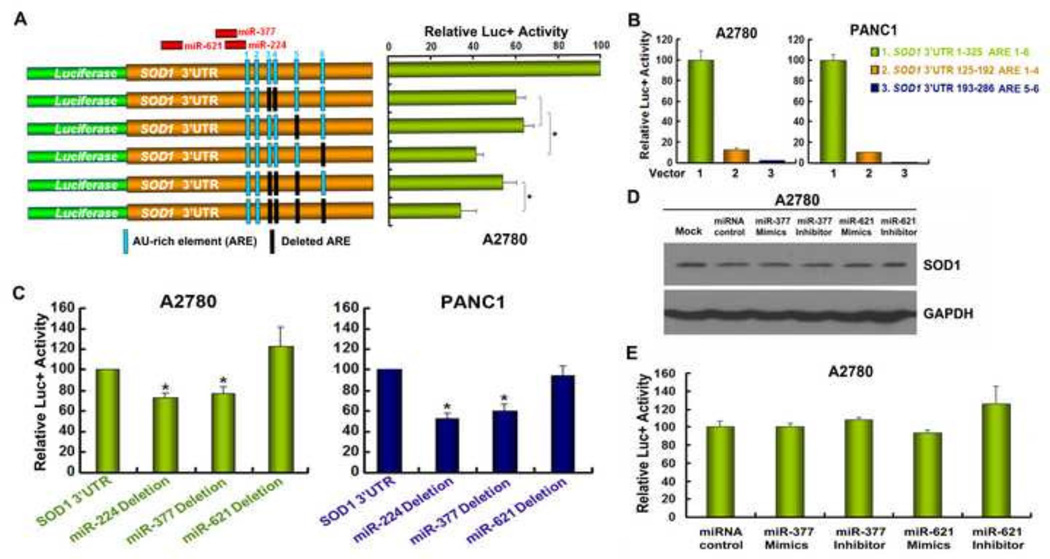
Figure 3. The integrity of the SOD1 3’UTR sequence is critical for maintaining SOD1 expression.
(A) A diagram indicates the relative positions of the 6 AREs (column) and miRNA binding sites. Various reporter constructs with deletion of the AREs (black column) or miRNA binding sites in the SOD1 3’UTR were constructed. (B) A2780 and PANC1 cells were transfected with the indicated constructs and assayed for luciferase activity after 48 h. (C) A2780 and PANC1 cells were transfected with SOD1 3’UTR reporter constructs with miR-224, miR-377 or miR-621 binding sites deleted. Luciferase activity was determined 48 h after transfection. (D) A2780 cells were transfected with miR-377 or miR-621 mimics/inhibitors and SOD1 expression was analyzed by Western blot 48 h post transfection. (E) pGL3-SOD1 3’UTR sense vector was co-transfected with miR-377 or miR-621 mimics/inhibitor, respectively and assayed for relative luciferase activities 48 h post tranfection. Data represent mean ± SEM of three independent experiments, * P < 0.05, compared with pGL3-SOD1 3’UTR sense vector-transfected cells.