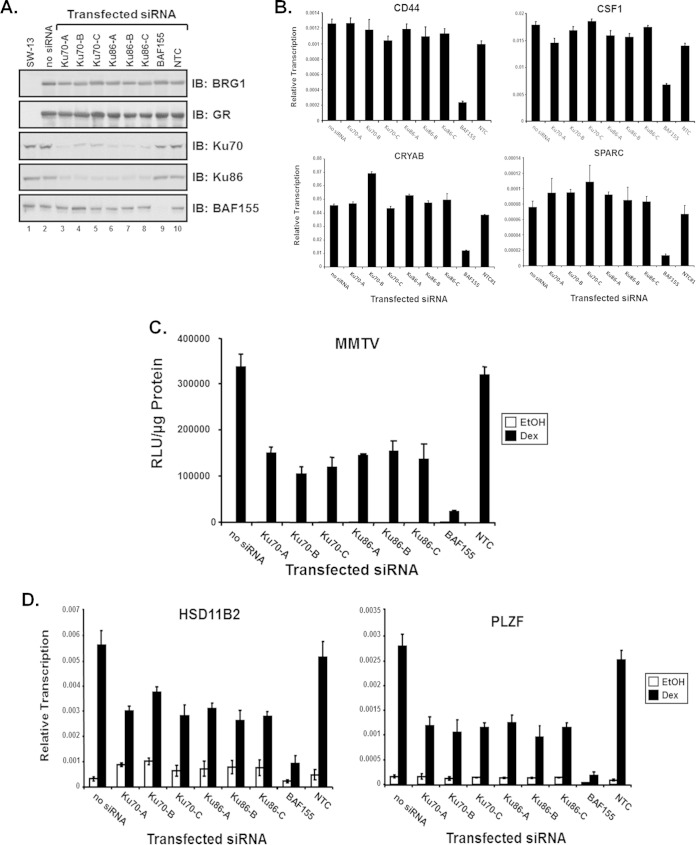
FIG 3.
Ku is required for GR-mediated BRG1-dependent transcriptional activation. Three siRNA duplexes specific for human Ku70 or Ku86 were used to evaluate the requirement of Ku in GR-mediated BRG1-dependent transcriptional activation of chromatin MMTV and endogenous genes. (A) Western blot analysis using total protein lysates from transfected SW-13/MMTV cells and antibodies specific for GR, BRG1, Ku70, Ku86, or BAF155 to monitor protein expression levels from transfected plasmids and to evaluate the extent of siRNA-induced protein knockdown. (B) Reverse transcription–real-time PCR analysis was used to determine the expression levels of endogenous BRG1-dependent genes in parental SW-13 cells cotransfected with BRG1 expression plasmid and the indicated siRNA duplexes specifically targeting Ku70, Ku86, or the core SWI/SNF subunit BAF155. Equal amounts of cDNA and gene-specific primers for CRYAB or SPARC were used for real-time PCR analysis. A nontargeting control (NTC) duplex siRNA used as a negative control. Quantitative analysis was performed with data normalized to GAPDH, and results are displayed as relative expression. Values are shown as mean ± standard deviation from 3 biological replicates. (C) SW-13 cells, containing a stably integrated MMTV reporter, were cotransfected with GR and BRG1 expression plasmids, and the indicated siRNAs were treated with 100 nM Dex or ethanol (EtOH) and assayed for relative luciferase activity. Relative MMTV luciferase activity was normalized to total protein measured and represented as relative light units (RLU). Values are shown as mean ± standard deviation (n = 3). (D) Reverse transcription-PCR analysis was used to determine the expression levels of endogenous GR/BRG1-dependent genes in SW-13 cells upon Ku siRNA protein knockdown. Total RNA extracted from treated SW-13 cells cotransfected with GR and BRG1 expression vectors and Ku70 or Ku86 specific siRNA duplexes was used as the template for cDNA synthesis. A nontargeting control (NTC) siRNA used as a negative control for protein knockdown. Values are shown as mean ± standard deviation from 3 biological replicates.