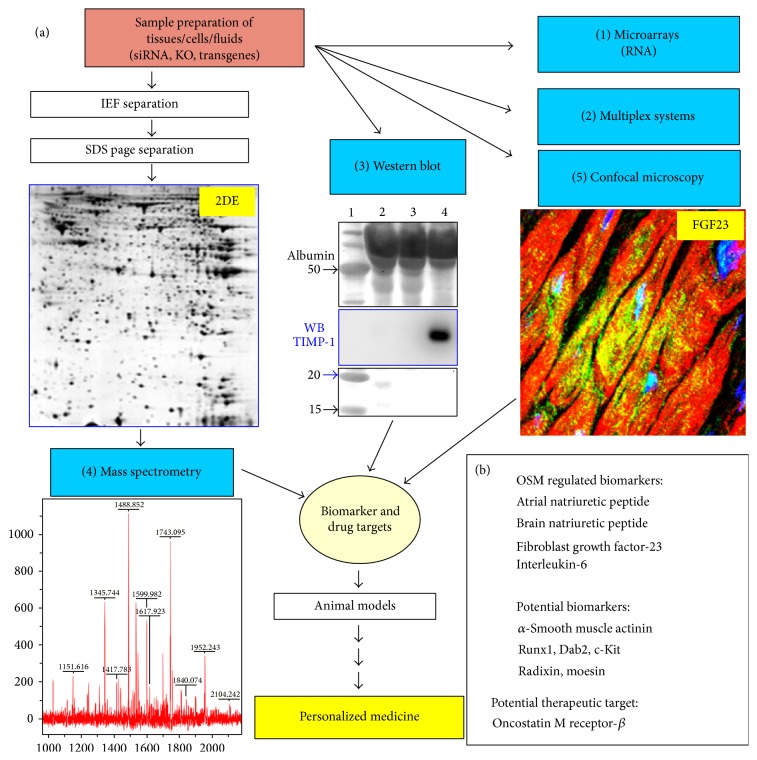
Figure 2.
The design of an “omics” technology research platform consisting of complementary core facilities for the future of a personalized medicine (adapted and modified from the “Venia legendi” work of J. Pöling, 2013). (a) Multiplex systems, Western blot, and confocal microscopy are antibody based methods. The Western blot shows a stained membrane resolving 1 μL of two serum samples (2&3) and 1 μL of pericardial fluid (PCF) from a patient with HF and high myocardial level of oncostatin M (size marker was run parallel in 1). A strong signal of TIMP-1 was detected in PCF after antibody hybridization of the membrane and subsequent chemiluminescence detection (WB). When targets are not known the separation of proteins lysates by 2-dimensional gel electrophoresis (2DE) reveals thousands of not yet defined protein spots after silver staining (proteome analysis). Here, a cytoplasmic cardiac 2DE of a 3-day-old rat is shown. Then, gels are scanned and compared and a computer based software program identifies regulated spots. (4) The protein spot is excised and identified by mass spectrometry combined with database searches. (1) In addition RNA might be extracted from the same sample and expression levels of ten thousands of genes can be simultaneously analyzed on DNA microarrays (transcriptome analysis). (5) Confocal microscopy verifies observations and provides further information about the protein. A confocal image shows FGF23-positive cardiomyocytes in a patient with aortic stenosis and high myocardial level of oncostatin M. (b) ANP, BNP, interleukin-6, and FGF-23 were identified as oncostatin M-regulated biomarkers on this platform. Further potential biomarkers such as radixin and moesin are indicated.