In 1996, Robinett et al. developed a way to visualize chromatin structure and dynamics in living cells.
In 1996, Robinett et al. developed a way to visualize chromatin structure and dynamics in living cells.
As a postdoc studying chromosomal structure with David Agard and John Sedat in the late 1980s, Andrew Belmont had observed intriguing ultrastructural details in interphase nuclei. “We’d see large-scale fibers about 100 nanometers thick, and then there’d be a 10- to 30-nanometer-thick fiber looping out, which we thought might represent an active gene,” says Belmont. “However, we couldn’t rule out that these features were simply artifacts of fixation or sample preparation because we couldn’t identify the region that we were looking at.”
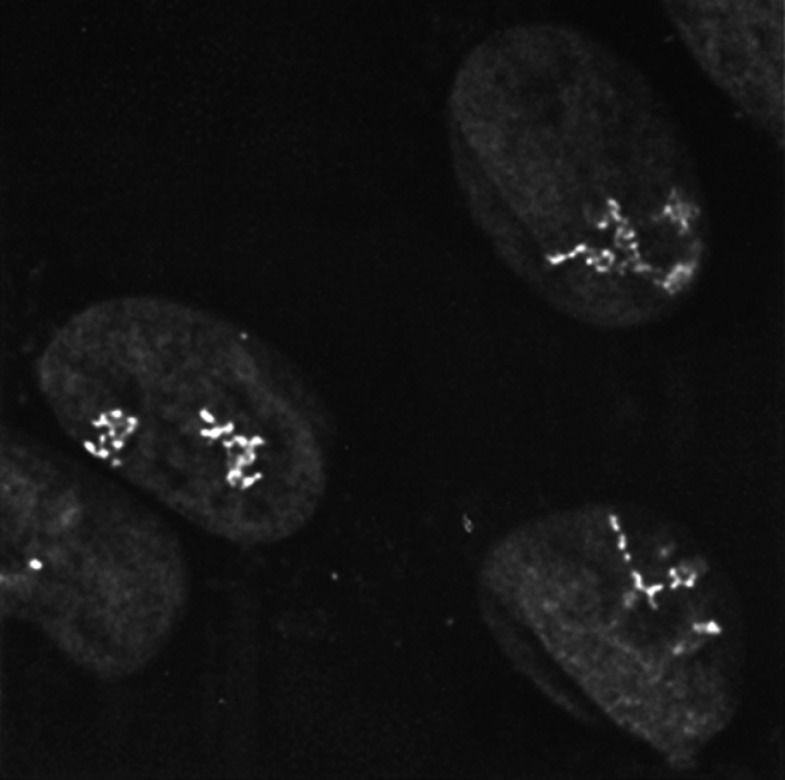
Lac repressor staining revealed distinct chromosomal architecture within different amplified chromosome regions.
ROBINETT ET AL., 1996
At the time, the only method available to identify a specific chromosomal region was in situ hybridization, in which a radioactive or fluorescently labeled nucleotide probe is annealed to complementary DNA or RNA sequences that have been denatured by chemicals or heat. The technique, first described in 1969 (1), enabled major advances in cytogenetics and genomic mapping (2). But it wasn’t suited to the application Belmont had in mind.
“One of the first experiments I did in my own lab was to perform a mock in situ hybridization and then take the sample to the electron microscope. I saw the chromatin structure was completely trashed at the ultrastructural level,” recalls Belmont. His group needed a way to label specific chromatin structures in cells that also preserved DNA ultrastructure. The approach they devised was described in a 1996 paper published in JCB (3).
“It seemed like a crazy idea at the time, and I was concerned it wouldn’t even work.”
The paper unveiled a genetic construct containing 256 lac operator repeats that, once integrated into a cell’s DNA, could be recognized by the lac repressor protein in both fixed and living cells. The site of lac repressor binding could then be visualized by indirect immunofluorescence or via fusion to the recently discovered green fluorescent protein. In mammalian cells, a single copy of the 256 repeat construct was sufficient to identify the site of its integration, which appeared under the light microscope as a tiny dot in the cell’s nucleus. Importantly, however, amplification of the inserted repeat also granted insight into chromosomal structure.
“We showed that different amplified regions fold in characteristic patterns,” notes Belmont. These patterns corresponded to whether the tag had integrated at open versus condensed chromosome regions. “We could observe large-scale chromatin fibers under a light microscope, then take them straight to the electron microscope and identify those fibers at the ultrastructural level by immunogold staining”—thus confirming that the large-scale chromatin fibers Belmont had previously seen in fixed extracted cells actually exist in live cells.
Finally, through collaboration with Andrew Murray’s lab, the researchers showed that it was possible to insert a lac operator tag at a specific site in the yeast genome. This made it possible to follow dynamics of specific chromosomal loci in living cells.
Their approach was successful, but Belmont admits he’s amazed it even got off the ground. “I kept outlining the project to prospective graduate students and no one would bite. It seemed like a crazy idea at the time, and I was concerned it wouldn’t even work.” It wasn’t clear the lac repressor would recognize the operator motif once it was assembled on nucleosomes, or that repressor binding would be detectable by the fluorescent probes and microscope cameras then available.
“It is often the case that ideas that look good on paper totally fail when confronted with the reality of biology,” agrees Carmen Robinett, first author on the paper. “Yet at every step, things just worked.”
Belmont says the credit for that goes to Robinett. “I didn’t even own a gel box at the time we began the work. I had no experience with any kind of recombinant DNA work, but Carmen basically came to the lab as a master’s student, and planned and executed what in retrospect was a really difficult cloning project to make the lac operator repeat construct. She generated the cell lines containing amplified chromosome regions with the inserts, and even established Drosophila lines carrying the repeats. But then she had to leave the lab to start her PhD degree in Berkeley, and the project languished for a year or more before my department head, Rick Horwitz, rescued me with funding for a technician.”
Belmont actually attributes his successful tenure application to this paper and the work it enabled, which includes investigations into fundamental questions about chromatin structure and localization (4, 5).
Others have found it quite useful as well. “Their proof-of-principle application was inspirational,” says former JCB Editor-in-Chief Tom Misteli. “It spawned an entire tool kit for tethering proteins to chromatin and for labeling, immobilizing and targeting chromatin regions. The system has generated insights into problems ranging from DNA repair and chromatin dynamics to gene positioning and nuclear body formation.”
References
- 1.Pardue, M.L., and Gall J.G.. 1969. Proc. Natl. Acad. Sci. USA. 64:600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volpi, E.V., and Bridger J.M.. 2008. Biotechniques. 45:385–409. [DOI] [PubMed] [Google Scholar]
- 3.Robinett, C.C., et al. 1996. J. Cell Biol. 135:1685–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belmont, A.S., et al. 2010. Cold Spring Harb. Symp. Quant. Biol. 75:453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khanna, N., Hu Y., and Belmont A.S.. 2014. Curr. Biol. 24:1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]