Visualization of single cadherins within cell membrane at nanometric resolution shows that E-cadherins arrange in ordered clusters and that these clusters control the anchoring of cadherin to actin and cell–cell contact fluidity.
Abstract
Oligomerization of cadherins could provide the stability to ensure tissue cohesion. Cadherins mediate cell–cell adhesion by forming trans-interactions. They form cis-interactions whose role could be essential to stabilize intercellular junctions by shifting cadherin clusters from a fluid to an ordered phase. However, no evidence has been provided so far for cadherin oligomerization in cellulo and for its impact on cell–cell contact stability. Visualizing single cadherins within cell membrane at a nanometric resolution, we show that E-cadherins arrange in ordered clusters, providing the first demonstration of the existence of oligomeric cadherins at cell–cell contacts. Studying the consequences of the disruption of the cis-interface, we show that it is not essential for adherens junction formation. Its disruption, however, increased the mobility of junctional E-cadherin. This destabilization strongly affected E-cadherin anchoring to actin and cell–cell rearrangement during collective cell migration, indicating that the formation of oligomeric clusters controls the anchoring of cadherin to actin and cell–cell contact fluidity.
Introduction
Around 35% of proteins in cells are in an oligomeric state (Goodsell and Olson, 2000). Oligomerization provides several functional advantages such as a mechanism to resist degradation and, more importantly, to make higher order long-living subcellular structures such as cytoskeletal filaments and functional nanomachines. Tissue cohesion is ensured by cell adhesion molecules that establish short living intercellular protein–protein bonds at the single molecule level (Perret et al., 2004). Oligomerization could provide the necessary strength to support intercellular adhesion and resistance to mechanical stress. Cadherins are major cell adhesion molecules in animal cells (Hulpiau et al., 2013). Cadherins diffusing at the plasma membrane initiate cell–cell interactions by establishing homophilic intercellular bonds (Mège et al., 2006). These trans-interactions analyzed by atomic force microscopy or biomembrane force probe have been shown to be short living (Baumgartner et al., 2000; Perret et al., 2004), implying that some higher order processes must take place for cadherin-mediated adhesion to reach sufficient stability to sustain physiologically relevant resistance to mechanical stress. Nascent cell–cell contacts initiated by cadherin trans-interactions evolve in adhesion plaques by the growth of cadherin clusters gathering additional trans-interacting cadherin molecules by a diffusion trapping mode (Adams et al., 1998; Lambert et al., 2007). Upon anchorage to the underlying actin cytoskeleton, which may bring additional cooperativity in cadherin recruitment as well as stability (Lambert et al., 2002; Hong et al., 2013), these adhesion plaques eventually evolve in adherens junctions (AJs; Mège et al., 2006). However, whether cadherin clusters found in AJs are organized in oligomeric structures as connexins in gap junctions (Raviola and Gilula, 1975) or desmosomal cadherins in desmosomes (Al-Amoudi et al., 2007), or have no particular organization as contradictorily reported for desmosomal cadherins (He et al., 2003), remains an open question.
Structural data have brought important information on the organization of cadherins (Overduin et al., 1995; Shapiro et al., 1995; Boggon et al., 2002; Shapiro and Weis, 2009). The current hypothesis is that adhesion starts with trans-interaction of EC1 domains of cadherins from apposed cells. More recently, a cis-interface for E-cadherin (Ecad) has been identified in crystal lattices. It involves the nonsymmetrical interaction of the EC1 domain of one cadherin with the EC2 domain of a neighboring cadherin (Harrison et al., 2011). Site-directed mutagenesis in EC1 (V81D) and EC2 (L175D) domains abolishes the formation of a cis-interface in the crystal without affecting the formation of the trans-interface. V81D, L175D-mutated Ecad ectodomain failed to form ordered junction-like structures in a liposome system, whereas wild-type (wt) Ecad did. Further theoretical and simulation work predicted that Ecad organizes in linear or more complex nanometric arrays as a result of trans- and cis-interactions (Wu et al., 2011, 2013). However, although Ecad cluster size and distribution have been reported with unprecedented resolution in tissues thanks to super-resolution microscopy (Truong Quang et al., 2013; Wu et al., 2015), cadherins have never been imaged at a nanometric resolution and thus no direct proof of ordered organization of cadherin in clusters has been provided so far in cells. Harrison et al. (2011) data suggest that the cis-interface stabilizes junctional Ecad. However, these data have been obtained by expressing wt and cis-Ecad forms deleted from the cytoplasmic domain. Because anchorage of cadherin cytoplasmic domain to actin via catenins is a major factor of AJ formation and strengthening (Lambert et al., 2002; Cavey et al., 2008; Hong et al., 2013), one may ask questions on the influence of cadherin oligomerization on cadherin cytoplasmic tail anchoring to F-actin.
The purpose of this work is to provide evidence for the predicted formation of arrays of oligomeric Ecad in cellulo and to study the functional implication of Ecad oligomerization on the formation and maturation of cell–cell contacts. We expressed full-length wt and cis-Ecad (V81D, L175D double mutant) in Ecad-deficient A431D cells and used an electron microscopy approach to visualize single Ecad molecules at the cell membrane. We also studied the consequences of the disruption of the cis-interface on cell–cell contact formation and stability. Our data provide evidence for an ordered organization of Ecad in clusters, depending on the cis-interface. We show that the cis-interface was, however, not required for AJ formation. The effects of the V81D, L175D mutations on Ecad turnover, association to catenin, and anchoring to actin were analyzed by cell imaging, FRAP analysis, and manipulation of Ecad-coated beads with magnetic tweezers. Ecad cis-oligomerization perturbations had moderate effects on Ecad complex stability but dramatically impaired Ecad anchoring to the actin cytoskeleton, the overall stability of cell–cell contacts, and collective cell behavior.
Results
Effect of the disruption of the cis-interface on Ecad expression and cell–cell adhesion
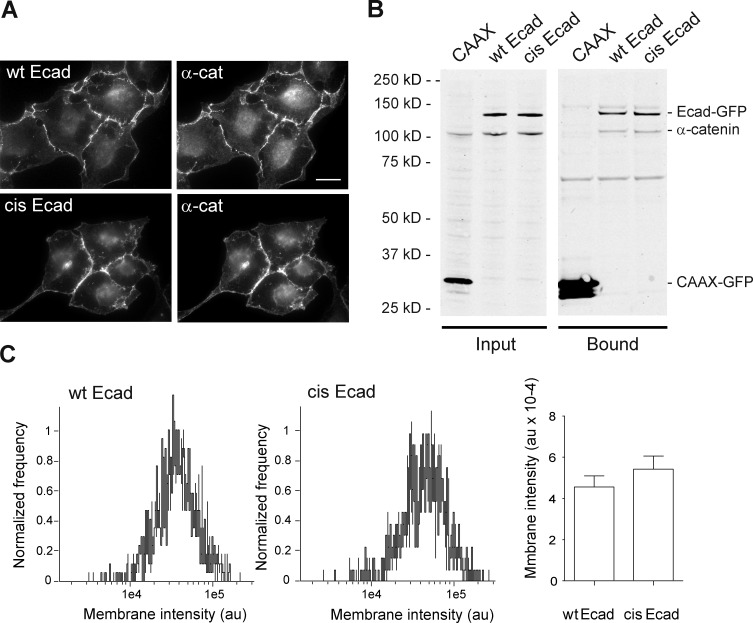
To visualize Ecad oligomers at the cell membrane, we used A431D cells, as they do not express Ecad (Lewis et al., 1997). Both wt Ecad-GFP and cis-Ecad-GFP were accumulated at cell–cell contacts where they recruited α- (Fig. 1) and β-catenin (Fig. S1, A and B) as previously reported (Troyanovsky et al., 2015). Western blot analysis indicates that wt Ecad-GFP and cis-Ecad-GFP were expressed at similar levels. Similar levels of α- and β-catenin were coimmunoprecipitated with the wt and cis-mutant Ecad, indicating that the impairment of Ecad cis dimerization did not affect the association to catenin. Cell surface fluorescence imaging indicates that cis-Ecad-GFP was as efficiently accumulated at the plasma membrane as wt Ecad-GFP and even slightly more (Fig. 1 C).
Figure 1.
wt Ecad and cis-Ecad expression restore Ecad-dependent cell–cell contacts in A431D cells. (A) Fluorescence imaging of cells expressing wt Ecad-GFP (wt Ecad) or cis-Ecad-GFP (cis-Ecad) reveals indistinguishable coaccumulation of Ecad and α-catenin at cell–cell contacts. Bar, 20 µm. (B) GFP-tagged proteins were immunoprecipitated from transfected cell lysates and subjected to Western blotting to detect GFP and α-catenin (Bound). Western blot of the cellular extracts before immunoprecipitation is shown on the left (Input). CAAX-GFP expressing cells were used as a control. Both wt Ecad-GFP and cis-Ecad-GFP were expressed at the predicted molecular mass (140 kD) and at similar levels (Input). α-Catenin was coimmunoprecipitated at similar levels with wt Ecad-GFP and cis-Ecad-GFP. (C) Representative distributions of cell surface–associated fluorescent intensities (arbitrary units [au]) for wt and cis-Ecad-GFP–transfected cells, 24 h after transfection (1,500 and 1,300 objects analyzed, respectively). The histogram represents the mean of the median fluorescent intensities ± SEM obtained from three independent experiments.
To test whether the impairment of cis dimerization alters cadherin-mediated cell adhesion, we measured the ability of Ecad-Fc–coated beads to bind to transfected cells (Fig. S2, C and D). No significant difference was seen in the number of Ecad-Fc coated beads bound on cis and wt Ecad expressing cells. These observations indicate that the disruption of the cis-interface did not affect the ability of Ecad to mediate cell adhesion, extending previous observations showing that the disruption of the cis-interface did not impair cell aggregation in the context of cells expressing tailless Ecad (Harrison et al., 2011).
In cellulo visualization of Ecad cis-oligomers
In an attempt to visualize cadherin oligomeric organization in cellulo, we used an electron microscopy approach, allowing to stoichiometrically label GFP-tagged proteins on membrane patches (Fig. 2 A). Cells were grown on Ecad-Fc–coated electron microscope grids, and then ripped off from the surface, exposing the cytoplasmic face of the plasma membranes. Membrane rip-offs were then incubated with gold nanoparticles (NP) functionalized at a 1:1 stoichiometry with anti-GFP-NP. NPs were then observed by transmission electron microscopy (TEM), allowing the visualization of single GFP-tagged cadherins. Few areas with very high density of anti-GFP nanobodies were observed for wt Ecad (up to 2698 NP/µm2) and were never observed for cis-Ecad (up to 279 NP/µm2; Fig. S2, A and E). Because wt and cis-Ecad were expressed at similar levels and only a tiny area can be visualized by TEM, this difference could be the consequence of differences in distribution of wt and cis-Ecad or of a greater propensity of cis-Ecad membrane to be teared off when scratching the cell roof from the Ecad-coated grid.
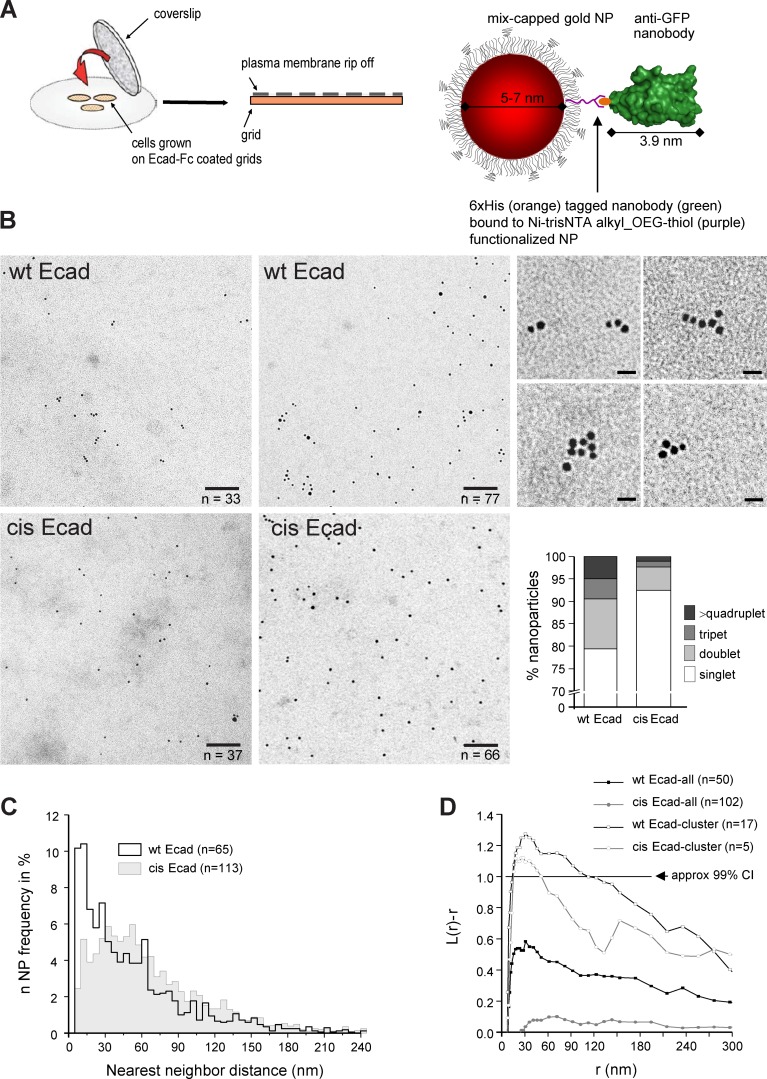
Figure 2.
wt Ecad-GFP molecules are organized in nanometric arrays at the cell membrane. (A) Experimental approach. (left) Transfected cells were seeded at low density on Ecad-Fc–coated grids. After 2 h, the upper part of the cells were ripped off, leaving plasma membrane sheets corresponding to the bottom part of the cells (inner leaflet face up) on the electron microscopy grids (adapted from Hancock and Prior, 2005). Grids were then incubated with 5–7-nm-diameter mix-coated gold NPs (in red) conjugated to a single anti–GFP-NB (in green) and observed by TEM. As a control of binding specificity we observed only a few anti-GFP-NP on plasma membrane of nontransfected cells (not depicted). (B) TEM visualization of anti–GFP-NP on plasma membrane sheets of wt Ecad-GFP (wt Ecad) and cis-Ecad-GFP (cis Ecad) cells spread on Ecad-Fc. The four images on the left are representative images for wt Ecad and cis-Ecad, with around 35 and 70 NPs, left and right, respectively. The exact number of NPs in the picture is given at the bottom right. On the right is close-ups of NP aggregates found in the wt Ecad condition. Percentages of monomers, doublets, triplets, and higher oligomeric forms are given in the bottom right graph. χ2 test shows significant difference between wt and cis-Ecad expressing cells regarding the proportion of singlets and oligomers (P < 0.01). Bars: (left) 100 nm; (right) 15 nm. (C) Distribution of the center to center distance between each NP and its nearest neighbor. n, number of images analyzed per condition; Kolmogorov-Smirnov non-parametric test reveals a significant difference between both distributions (P = 0.006). (D) NP clustering for wt Ecad (black) and cis-Ecad (gray) expressing cells was characterized by K-function analysis. Curves with solid symbols correspond to the mean L(r) − r values calculated for all images. Curves with open symbols correspond to the mean L(r) − r values calculated for individual images presenting significant clustering. Values of L(r) − r above the 99% confidence interval (CI; black line) indicate significant clustering. Proportion of images having clustering of NP for wt and cis-Ecad is significantly different according to χ2 test (P < 0.01).
To further compare the NP distribution between wt and cis-Ecad expressing cells only fields having a similar density of labeling (4–180 NP/µm2) were considered for the analysis (Fig. 2 and Fig. S2, B and E). A fraction of NP-bound wt Ecad was distributed in doublets, triplets, or lines of a few particles as well as in small ordered clusters (Fig. 2 B and Fig. S2 C). Some aggregates of NP were seen for cis-Ecad but to a much lower extent. Analysis of the distance between each NP and its nearest neighbor revealed that 20.6% of the NPs were at <15-nm distance (Fig. 2 C). For this fraction, the mean center to center distance of neighboring particles was 10.1 ± 2.4 nm with a pick at 8 nm (Fig. S2 F); thus, in the range of the predicted distance between two adjacent cadherins interacting in cis (7.2 nm; Harrison et al., 2011). The proportion of oligomers and distance distribution were significantly different for cis-Ecad with only 7.6% of the NPs at <15 nm, a peak in the distance distribution between 30 to 55 nm, and very few NPs with a distance <10 nm (Fig. 2 C and Fig. S2 F). To determine whether the oligomeric forms observed reflect significant clustering, Ripley’s K-function was used (Prior et al., 2003). 34% of the images acquired for wt Ecad cells present a significant clustering within a 15- to 120-nm range, with a maximum deviation out of the 99% confidence interval occurring at a radius of 30 nm (Fig. 2 D). In contrast, only 4.9% of the images acquired for cis-Ecad cells present a significant clustering within a 15- to 51-nm range, with a maximum deviation out of the 99% confidence interval occurring at a radius of 30 nm. cis-Ecad molecules were more randomly distributed, indicating that the cis-interface largely participates in the clustering of Ecad molecules. Altogether, these data provide supporting evidence that Ecads engaged in trans-interactions organize in ordered oligomers that require a proper cis-interface. In addition, the observed organization of particles within clusters, in particular in higher density images (Fig. S2 D), was highly reminiscent of the one predicted by theoretical models (Wu et al., 2011, 2013). This is, to our knowledge, the first time nanometric distribution of Ecad molecules can be visualized in a cellular context.
AJs form independently of Ecad cis-oligomerization
We then asked whether the disruption of this interface would affect the formation of AJs, which are believed to result from clustering of Ecads in restricted domains of cell–cell contacts (Mege et al., 1988; Harrison et al., 2011). We analyzed the propensity of transfected cells to organize Ecad in discrete cadherin adhesions when spread on Ecad-Fc, as a proxy of the ability of cells to form AJs (Gavard et al., 2004b). Both cis-Ecad and wt Ecad expressing cells spread on Ecad-Fc, and wt Ecad and cis-Ecad molecules were similarly recruited in radial cadherin adhesions, colocalizing with actin fibers (Fig. 3 A), suggesting that the formation of AJs may not involve the cis-interface.
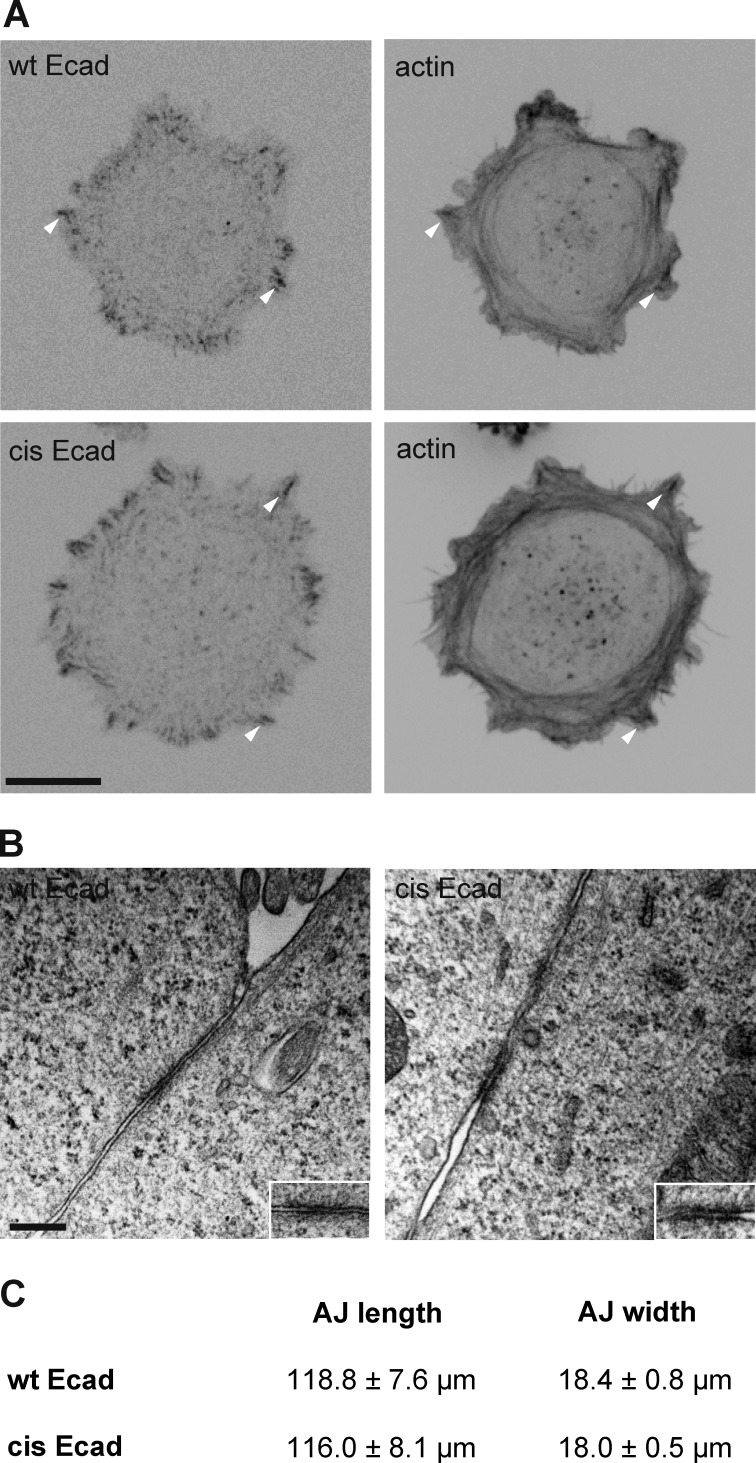
Figure 3.
Ecad cis-oligomerization is not required for AJ formation. (A) wt Ecad-GFP (wt Ecad) and cis-Ecad-GFP (cis Ecad) cells coexpressing LifeAct-Ruby were fixed after 2 h of spreading on Ecad-Fc–coated surfaces and imaged for Ecad-GFP and F-actin. Bar, 20 µm. The disruption of the cis-interface did not impact the ability of transfected cells to spread on Ecad-Fc and to recruit Ecad in cadherin adhesions (arrowheads). (B) Transmission electron micrographs showing the ultrastructure of intercellular contacts of wt Ecad-GFP (wt Ecad) and cis-Ecad-GFP (cis-Ecad) expressing cells. Bar, 100 nm. Insets show the junctional areas. (C) Table showing the mean length (± SEM) and the mean width (intermembrane distance; ± SEM) of AJs formed by wt Ecad (n = 36) and cis-Ecad-GFP expressing cells (n = 24). The disruption of the cis-interface does not prevent the formation of AJ.
To directly investigate the ability of cis-mutant proteins to induce the formation of AJs, we performed an ultrastructural examination of transfected cell monolayers by TEM. We searched for the presence of AJs, defined as zones of straight membrane apposition associated with the presence of dense material (Perez-Moreno et al., 2003). As reported by Lewis et al. (1997), A431D cells formed neither desmosomes nor AJs (unpublished data), allowing unambiguous interpretation of the observations on wt Ecad and cis-Ecad expressing A431D cells. The expression of wt Ecad drastically changed the cell–cell contact ultrastructure with the appearance of AJs with a mean intermembrane distance of 18.4 ± 0.7 nm (Fig. 3 B). Typically, cis-Ecad cells presented indistinguishable intercellular junctions with similar mean intermembrane spacing (18.0 ± 0.5 nm). Moreover, the mean lengths of these structures were similar. Altogether, TEM analysis demonstrates that the cis-interface is not required for the formation of AJs.
Ecad cis-oligomerization stabilizes junctional cadherins
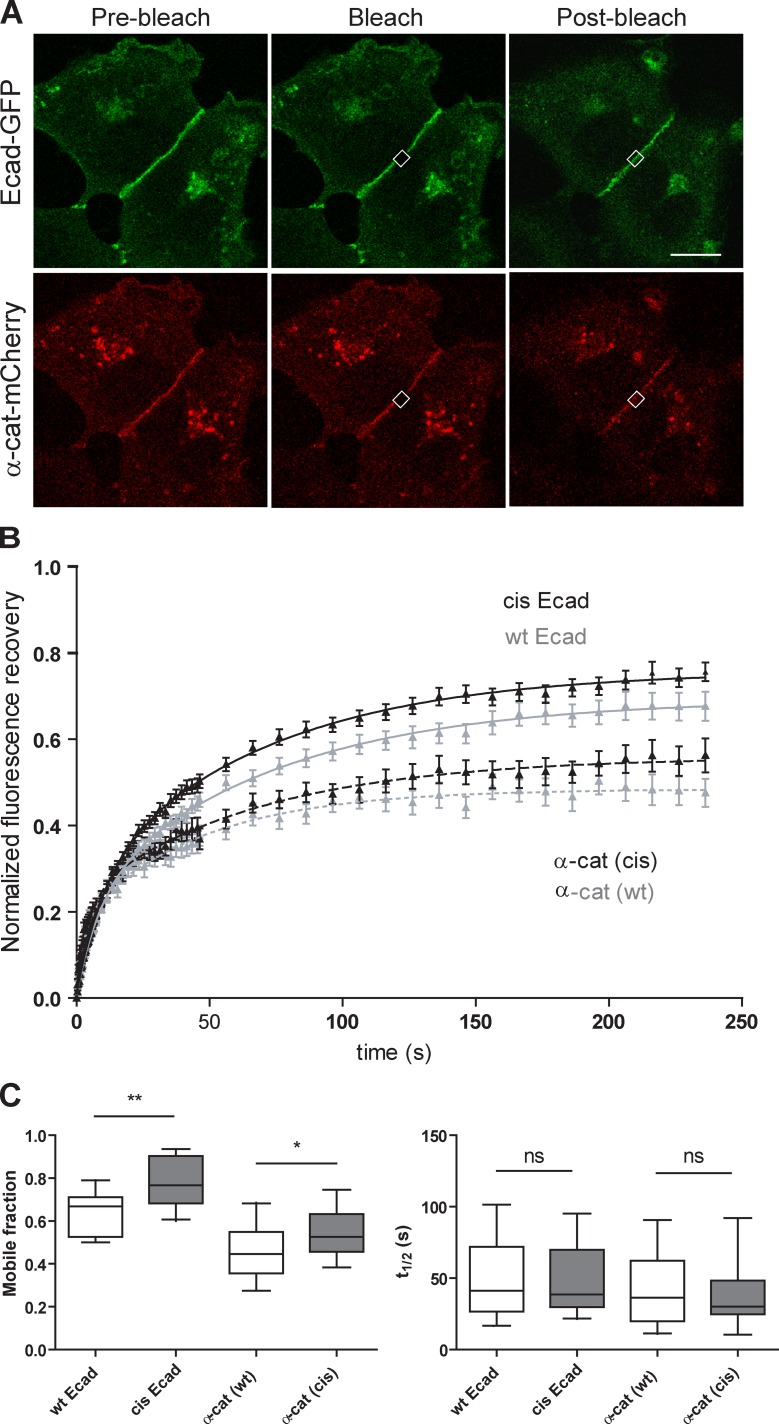
We hypothesize that the cis-interface may, however, affect the dynamics of junctional molecules. To test whether mutations affecting the cis-interface had an effect on full-length Ecad dynamics, we performed FRAP experiments on wt and cis-Ecad expressing cells (Fig. 4, A and B). The disruption of the cis-interface induced a moderate but significant increase of the mobile fraction of Ecad. The t1/2 value was not affected (Fig. 4 C), suggesting that the mobility of fast diffusing molecules was not altered. To have access to the dynamics of the cytoplasmic partners of Ecad, dual wavelength FRAP was performed on cells coexpressing α-catenin-mCherry. Impairment of cis-interactions induced similar trends in the dynamics of α-catenin (Fig. 4, B and C). The mobile fraction of α-catenin at cell–cell contacts significantly increased, whereas the t1/2 was not affected. Altogether, these observations are in good agreement with a contribution of the cis-interface, through the formation of ordered oligomeric structures, in stabilizing junctional Ecad. The impairment of the Ecad cis-interface led to a proportional increase of the α-catenin mobile fraction, suggesting that the increase in cadherin stability brought by ectodomain oligomerization has a direct intracellular impact through modifications of α-catenin dynamics. This may in turn affect cadherin anchoring to actin filaments. Such moderate changes in cadherin dynamics are expected to have limited impact on the steady-state recruitment of cadherin and catenin molecules and formation of AJs in agreement with photon end electron microscopy observations.
Figure 4.
Ecad cis-oligomerization stabilizes cadherin–catenin complexes at cell–cell contacts. (A) Characteristic images of GFP and mCherry signal before (Pre-bleach), immediately after bleaching (Bleach), and 250 s after the bleach (Post-bleach) performed on wt Ecad-GFP and α-catenin-mCherry doubly transfected cells. White squares represent the bleached region. Bar, 20 µm. (B) Normalized fluorescence recovery curves versus time for wt Ecad-GFP, cis-Ecad-GFP, and α-catenin-mCherry in wt Ecad and cis-Ecad expressing cells (n ≥ 29). (C) Box and whiskers plots (median + 10–90%) showing the mobile fraction (left) and the t1/2 (right) extracted from a one-exponential decay fit of fluorescence recovery curves. The disruption of the cis-interface led to a moderate increase of the mobile fraction of Ecad and α-catenin molecules without apparent modification of the diffusion characteristic times. *, P < 0.02; **, P < 0.05; ns, not significant (paired Student’s t test; n ≥ 23).
Ecad cis-oligomerization stiffens the mechanical coupling of cadherin adhesions to actin
The functional anchorage of cadherin–catenin complexes to F-actin has emerged as a major signaling pathway downstream of cadherins (Giannone et al., 2009; Takeichi, 2014), acting on the reinforcement of cell–cell contacts (le Duc et al., 2010; Yonemura et al., 2010; Thomas et al., 2013). To compare the functional anchorage of wt Ecad and cis-Ecad to actin we first studied actin dynamics in the lamellipodia of cells spread on Ecad-Fc–coated surfaces. Indeed, according to Mitchison and Kirschner (1988), a decreased actin treadmilling speed correlates with an increased friction between the cytoskeleton flowing underneath the plasma membrane and the membrane-anchored adhesion sites. Actin retrograde flow was visualized by live-cell imaging, thanks to the coexpression of LifeAct-Ruby (Fig. 5 A and Videos 1 and 2). The speed of the rearward flow of actin was increased by 30% in cis-Ecad compared with wt Ecad expressing cells, suggesting that cadherin oligomerization is involved in the coupling of the actin cytoskeleton to the plasma membrane. Thus, the increase in actin retrograde flow observed when the Ecad cis-interface was disrupted reflects a reduced association of Ecad clusters to the actin network. This increase in actin dynamics was correlated with an increase in cell protrusion (Fig. 5 B). Both the maximum amplitude and the frequency of lamellipodia edge back and forth movements were increased when the cis-interface was mutated (Fig. 5 C). These increases in actin rearward flow and protrusion activity in the lamellipodia of cells spread on Ecad-Fc are also suggestive of a decreased anchoring of cis-Ecad clusters to actin.
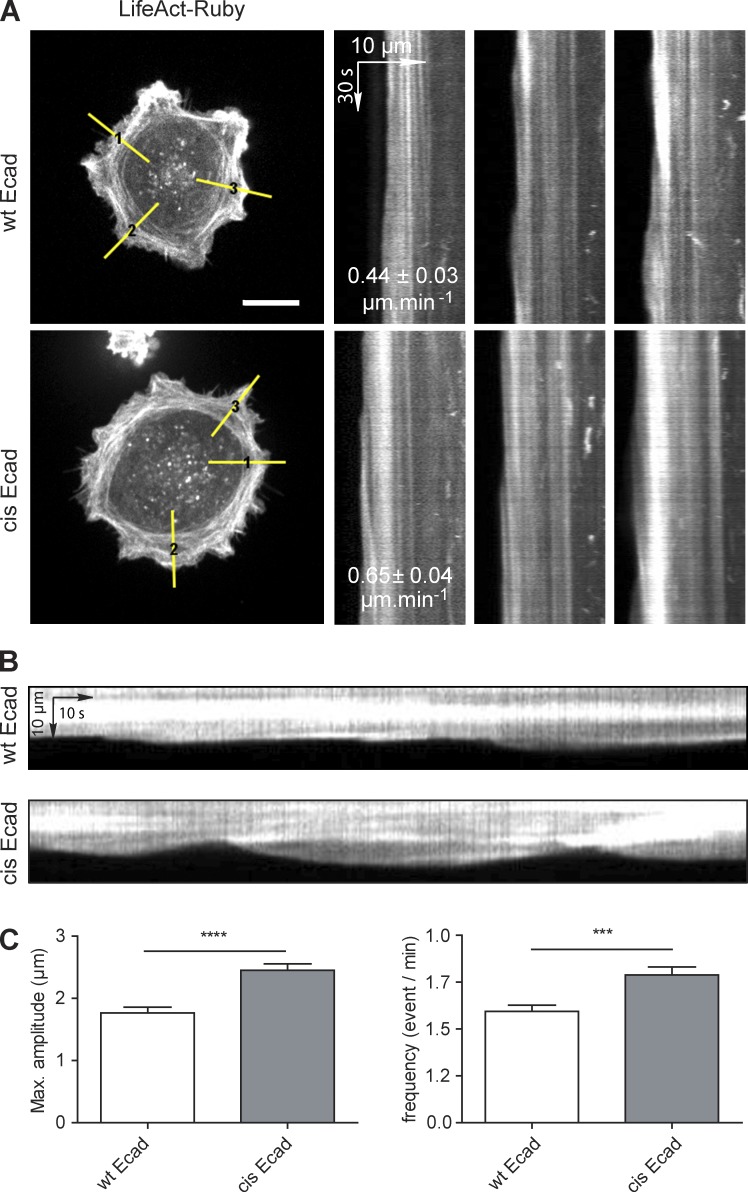
Figure 5.
Impairment of Ecad cis dimerization alters the actin dynamics of cells spread on Ecad-Fc. Cells coexpressing wt Ecad-GFP or cis-Ecad-GFP and LifeAct-Ruby were seeded on Ecad-Fc substrates for 2 h and then subjected to spinning disk live-cell imaging for 3 min at a frequency of one image per 500 ms. (A) Still Images of LifeAct-Ruby distribution. Bar, 25 µm. The actin retrograde flow was quantified by kymograph analysis (yellow lines 1–3, 1 pixel width, perpendicular to the cell membrane in Ecad dense region). Superimposed on the kymographs are the means of actin retrograde flow speed for wt Ecad (n = 156 kymographs from 26 cells) and cis-Ecad (n = 192 kymographs from 32 cells) cells. The actin retrograde flow was significantly faster for cis-Ecad expressing cells than for cells expressing wt Ecad (P ≤ 0.0002, Student’s t test). (B) Similar kymographs of the LifeAct-Ruby signal extending on a longer time window revealed the cyclic protrusion of the edge of wt Ecad and cis-Ecad expressing cells. (C) Quantification of the maximum amplitude and frequency of membrane protrusions (mean values ± SEM; n = 100 kymographs from 26 cells for wt Ecad cells and n = 130 kymographs from 32 cells for cis-Ecad expressing cells). ****, P ≤ 0.0001; ***, P ≤ 0.005, Student’s t test.
To directly test the mechanical coupling of wt and cis-Ecad to the underlying cytoskeleton, we probed the response to force of Ecad-coated magnetic beads bound to wt and cis-Ecad cells, using magnetic tweezers (Kollmannsberger and Fabry, 2007). We generated a magnetic field in the vicinity of beads applying forces ∼20 pN (Fig. 6 A). The semi-quantitative analysis of bead behavior as the magnetic power was turned on indicates that beads adhered less firmly to cis-Ecad expressing cells than to wt Ecad expressing cells (Fig. 6 B). Single beads bound to wt Ecad cells and cis-Ecad cells were then subjected to successive steps of force application (Fig. 6 C and Videos 3 and 4). No clear trend in the changes of the displacement amplitude in function of the number of cycles performed was noticed, suggesting that bead–cell mechanical coupling was not subject to force-dependent reinforcement as previously reported (Lambert et al., 2002). However, the amplitude of bead displacement was consistently higher for Ecad-Fc beads attached to cis-Ecad cells than for those bound to wt Ecad cells (Fig. 6 D). Altogether, these data indicate that Ecad cis-oligomerization increases the stiffness of Ecad complex coupling to internal structures.
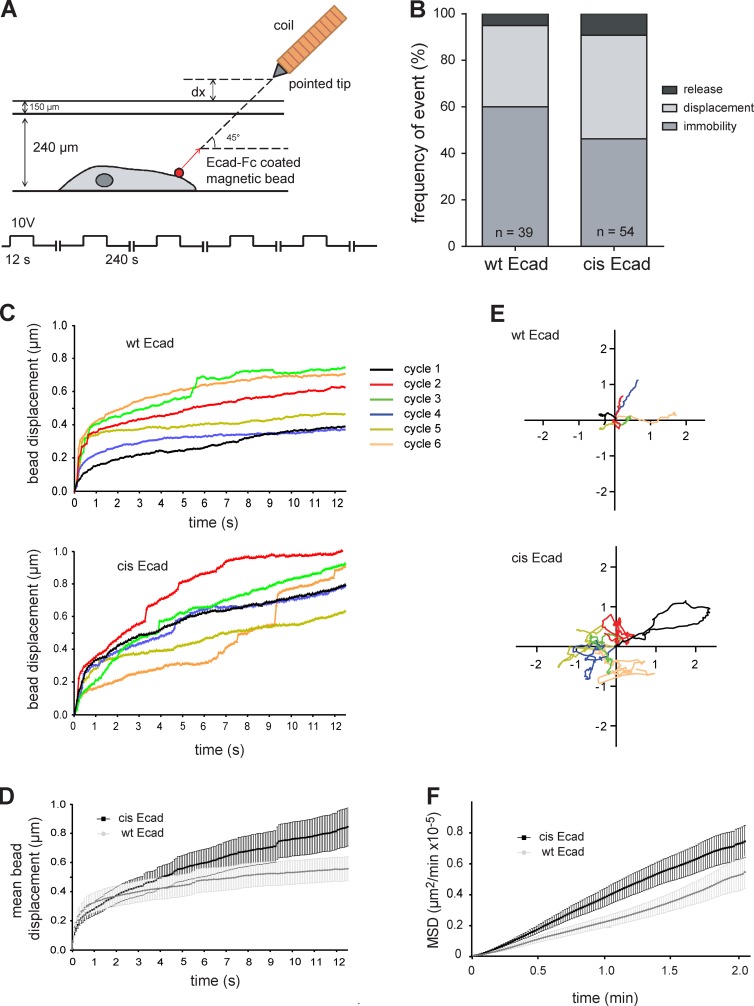
Figure 6.
Ecad oligomerization increases the anchoring of cadherin adhesions. (A) Magnetic tweezers experimental setup. A pointed iron tip is wrapped with a copper coil under tension to generate a magnetic field in the vicinity of beads. 2.8 µm of Ecad-Fc–coated magnetic beads were preincubated 1 h on wt Ecad or cis-Ecad expressing A431D cells, and then the unbound beads were washed away. A 10-V magnetic field was applied during 10 s in the vicinity of a bound bead, and then the magnetic power was turn off for 240 s while acquiring phase-contrast images to follow bead displacement. Beads were tracked during and in between the application of forces. This sequence was repeated six times over each analyzed bead. (B) Distribution in three classes (release, displacement, and immobility) of the responses to the magnetic field of Ecad-Fc–coated beads bound to wt Ecad (39 cells) and cis-Ecad (54 cells) expressing cells. (C) Bead displacement under force: representative traces of the displacement from origin of single beads bound to wt Ecad cells and cis-Ecad cells, respectively, in response to six successive cycles of magnetic force application. (D) Curves showing the mean displacement from the origin in response to forces of beads bound to wt Ecad cells (77 displacements measured on 12 independent beads) and cis-Ecad cells (72 displacements measured on 12 independent beads). (E) Bead displacement under zero force: successive trajectories (125 s long) undergone by a single bead bound to wt Ecad and cis-Ecad expressing cells, respectively, during the six successive steps of force release. Data shown are representative of the behavior of 12 beads for each condition. (F) MSD in the absence of force calculated over the six cycles for n = 12 beads attached to wt Ecad (gray) and cis-Ecad (black) expressing cells.
We also took advantage of this experimental setup to extract information on the mobility of wt Ecad and cis-Ecad molecules in the cell membrane, which has been demonstrated to reflect anchoring of the molecules to the actin cytoskeleton (Lambert et al., 2002). Fig. 6 E shows successive x-y trajectories undergone by single beads bound to wt Ecad and cis-Ecad expressing cells in the absence of force. Beads bound on cis-Ecad expressing cells moved over larger areas than those bound on wt Ecad expressing cells, revealing a weaker anchorage to the underlying cytoskeleton. Extraction of mean square displacement (MSD) revealed a significant difference in the displacement of the probed Ecad forms, with cis-Ecad being more mobile than wt Ecad molecules (Fig. 6 F). These data demonstrate that disruption of the cis-interface increases Ecad mobility. They indicate that the formation of ordered clusters allowed by the cis-interface strongly regulates the anchoring of cadherins to the actin cytoskeleton, resulting in the stiffening of the cadherin–actin mechanical link.
Disruption of the cis-interface strongly impacts collective cell migration
Although wt Ecad and cis-Ecad cells did not display differences in the ability to form cell–cell contact at confluence, we postulated that the formation of structured clusters by its action on the strengthening of the Ecad–actin mechanical link may have an impact on the stability of cadherin adhesions, which may be revealed only when cell–cell adhesions are challenged, for example, during collective cell movement. Indeed, previous studies showed that changes in intercellular adhesion had a strong impact on collective cell migration behaviors (Petitjean et al., 2010; Tambe et al., 2011; Vedula et al., 2014). To unravel the impact of Ecad cis-interface disruption, we performed a cell layer expansion assay after release of confinement (Fig. S3 A). When confluence was reached the block was removed, freeing space for cell layer expansion. The migration front was followed over 24 h and analyzed for its progression and roughness (Fig. 7 A). Both wt and cis-Ecad cell monolayers expended linearly. However, this displacement was 1.6 times faster in the case of cis-Ecad cells. The roughness of the migration front increased during the first 12 h as front cells moved toward the freed area and then stabilized at a plateau. However, the roughness at the front of cis-Ecad expressing cell layers both increased more quickly and reached a higher value at plateau (Fig. 7 A). This was because of individual cis-Ecad expressing cells that tend to escape the monolayer. In addition, some cis-Ecad cells transiently detached from the monolayer, in such a way that has never been observed with wt Ecad expressing cells (Videos 5 and 6).
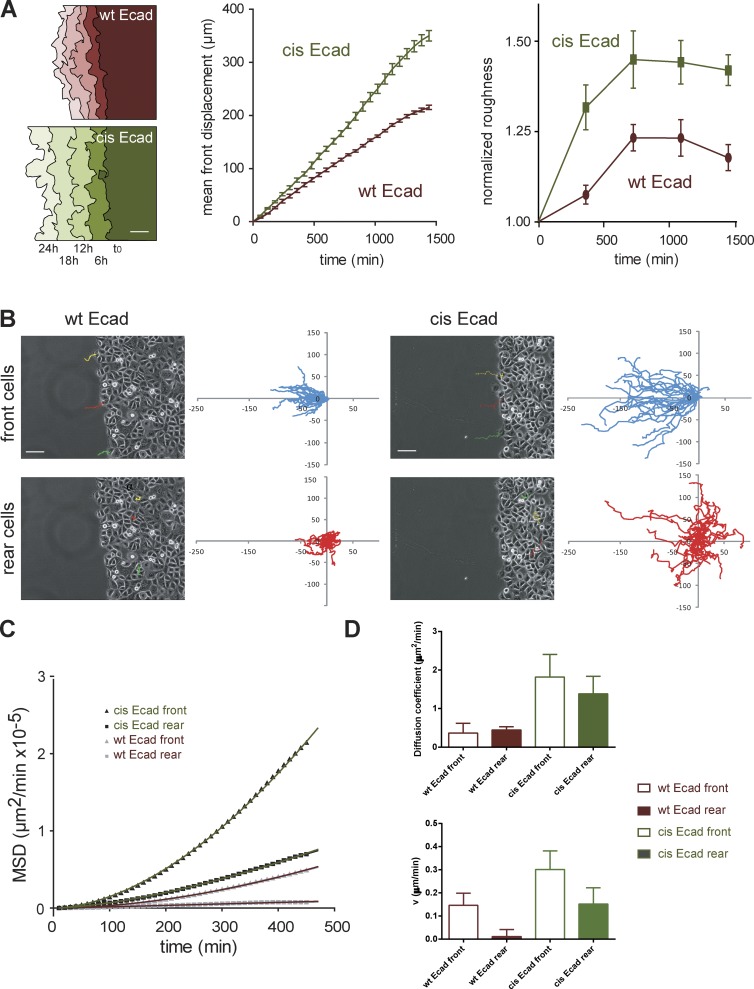
Figure 7.
The disruption of Ecad cis-oligomerization impairs collective cell migration. Cells were phase-contrast imaged starting at the time the PDMS block was removed (t0) and for 24 h. (A, left) Examples of the evolution of the migration front in function of time for wt Ecad and cis-Ecad expressing cells. Bar, 50 µm. Plots show the front migration displacement in function of time (middle) and the normalized front roughness in function of time (right). n = 12 and 17 for wt Ecad and cis-Ecad expressing cells, respectively. (B) Single cell tracking was performed over the first 8 h of the 24-h movies. Phase-contrast images of wt Ecad (left) and cis-Ecad (right) expressing monolayers taken at t0 with superimposed 8-h trajectories of single cells at the front and rear (cells of the fifth row away from the edge). Bars, 50 µm. Plots of 8-h trajectories of front (blue curves) and rear (red curves) cells for wt Ecad and cis-Ecad expressing monolayers. Axes are scaled in micrometers; n = 48 and 36 trajectories for wt Ecad and cis-Ecad expressing cells, respectively. cis-Ecad cells migrate on significant larger distances than wt Ecad cells both at the front and rear. (C) MSD as a function of time for trajectories presented in B and fits by the equation (MSD = 4Dt2 + v2t2). D: Histograms showing the D and v values (± SEM) extracted from the fits for wt Ecad and cis-Ecad front and rear cells.
This analysis suggests that cis-Ecad expressing cells migrate faster as a result of reduced cell–cell cohesion releasing the constraints imposed by neighboring cells. However, faster progression of the migration front might also result from increased cell autonomous migratory behavior. We thus analyzed the migration of isolated wt Ecad and cis-Ecad cells (Fig. S4). Both cell types displayed comparable individual cell behavior, indicating that the faster migration of cis-Ecad cells was a result of collective cell behavior. Alternatively, during the extension of the monolayer, numerous divisions were taking place, suggesting that changes in the division rate might directly have an impact on monolayer front progression. To put aside this possibility, we verified that occurrence of mitosis in the extending monolayer was not different for wt Ecad and cis-Ecad cells (Fig. S5 A). In addition, 5-ethynyl-2-deoxyuridine incorporation showed that there was no difference in the proportion of cells in S phase in each population (Fig. S5 B). Thus the differences in the migratory behavior do not result from altered cell autonomous properties but is specific of collective behavior.
To further support this hypothesis we analyzed the trajectories of cells located either at the migration front or in the back (at least at the fourth row). For both cell types, individual cells at the front had very directional trajectory projecting for the majority in a 40–50° angle cone perpendicular to the migration front. In contrast, cells at the back had more randomly oriented trajectories (Fig. 7 B). However, cis-Ecad cells covered much larger distances both at the front and rear. cis-Ecad cells migrated almost two times faster than wt Ecad cells both at the front and rear (Fig. S3 B). To further describe the migration behavior of these cells, we extracted MSD values from individual cell trajectories (Fig. 7 C). The evolution of MSD as a function of time further showed that cells with the largest displacement were front cis-Ecad cells followed by rear cis-Ecad, and then front wt Ecad and rear wt Ecad cells. MSD curves were fitted using the equation MSD = 4Dt + v2t2, where 4Dt represents the Brownian motion component and v2t2 represents the directed motion component (Fig. 7 D). Diffusion coefficent (D) value for cis-Ecad cells both at the front and rear were significantly higher than the ones of wt Ecad cells, revealing the increased exploratory behavior of the cis-mutant cells versus their wt counterparts. This can be interpreted as an increased fluidity of the cis-Ecad monolayers caused by a higher instability of cell–cell contacts and an increased ability of cells to exchange partners. The comparison of the second term of the equation (v: velocity of the directed movement) shows that front cells of both phenotypes display a more directed motion than the cells at the rear, which is expected because these cells face an empty space. However, the directed motion velocity of cis-Ecad cells is twice higher than the one of wt cells. Altogether these results suggest that the migration of wt Ecad cells is restrained by stable cell–cell contacts formed between neighboring cells within the monolayer. These results thus demonstrate that the stabilization of cell–cell contacts brought by the formation of the cis-oligomers is essential for coordinated cell behavior during collective cell migration.
Disruption of the cis-interface strongly reduces cell–cell coordination
To confirm that cell–cell coordination was affected by cis-interface impairment, we further analyzed the dynamic properties of expanding wt Ecad and cis-Ecad cell monolayers by particle image velocimetry (PIV; Fig. 8). PIV has been used as a powerful tool for quantitative analysis of tissue fluidity resulting from cell–cell rearrangements during collective cell migration (Vedula et al., 2012; Doxzen et al., 2013). PIV analysis confirmed that the mean instantaneous migration speed of cis-Ecad expressing cells was twice the one of wt Ecad cells (Fig. 8, A and B). From the velocity fields we calculated an order parameter as well as a correlation length. The order parameter reflects the degree of orientation of the velocity field in respect to a given direction, which is here defined as perpendicular to the initial migrating front (Fig. 8 C). Order parameter was maximum at the front but significantly lower for cis-Ecad than for wt Ecad cell layers. This order parameter decreased from the front toward the rear of the expanding monolayer. However, it was maintained at high values deeper in the monolayer for wt Ecad cells than for cis-Ecad cells. Finally, the correlation length, reflecting the mean distance at which velocity vectors are maintained in the same orientation, was also decreased when the cis-interface was impaired (Fig. 8 D). This parameter reflects the length over which cells are mechanically coupled, which is in the order of two to three cells for wt Ecad cells and only one cell for cis-Ecad cells. Altogether these observations show that cis-Ecad cell monolayers behave as a more fluid material, implying more rearrangement between individual cells. In agreement with single cell tracking and front displacement analysis, these observations demonstrate that when the cis-interface was impaired the apparent fluidity of the cell monolayer was increased as a direct consequence of decreased cell–cell cohesion, allowing more cell–cell rearrangements.
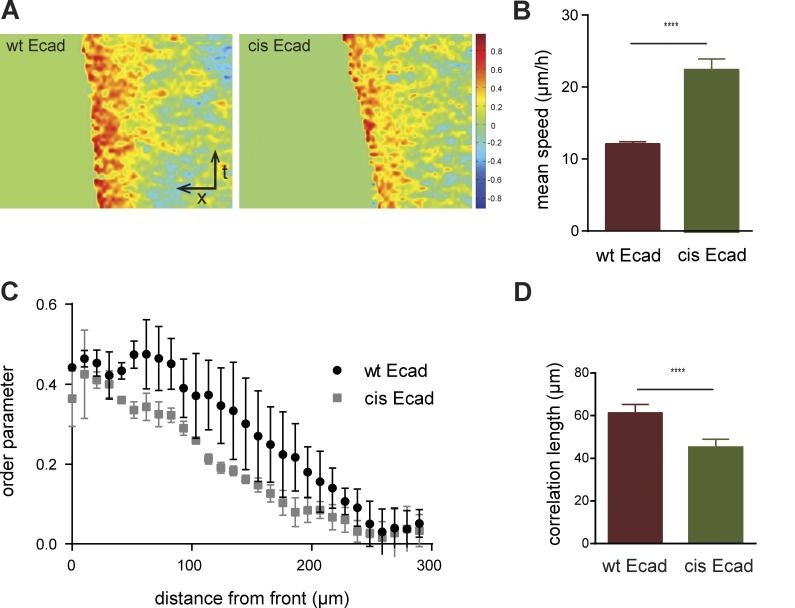
Figure 8.
PIV analysis of wt Ecad and cis-Ecad cell migration. (A) Instantaneous velocities were extracted 250–300-µm deep from the migration front for each image and spatially averaged along the migrating axis in kymographs, giving heat maps of the order parameter (−1 means backward and +1 means forward movements). Mean instantaneous velocities (B), order parameters in the velocity field as a function of distance to the front (C), and mean correlation length (D) were extracted from the kymographs (± SEM). ****, P ≤ 0.0001, Student’s t test. wt Ecad cells displayed lower migration speed, migrate forward in a more directed fashion at the monolayer expansion front, and show better correlation in their movements.
Discussion
It is proposed that oligomerization of cell adhesion molecules and their association to the underlying cytoskeleton provide to intercellular junctions the necessary strength to ensure tissue cohesion. To address this hypothesis, we determined the nanometric organization of Ecad in the plasma membrane and studied the consequences of the disruption of the cis-interface on single molecule organization up to the multicellular scale. We provide the first description of the nanometric distribution of Ecad in cellulo. Surprisingly, Ecad cis-interface is not required for AJ formation. However, its mutation strongly impaired the mechanical coupling of adhesion complexes to actin filaments affecting cell–cell contact strength. This reduced linkage to actin strongly affects cell movement coordination, leading to increased cell migration. Altogether, we show that Ecad oligomerization occurs in a cellular context and we provide direct evidence that it participates in mechanical anchoring of cadherin clusters to the cytoskeleton.
We used NP labeling to reveal an ordered nanometric organization of Ecad molecules at the cell membrane. The minimal distance between two Ecads was 8 nm. However, the mean distance (10.1 nm) between two adjacent Ecads is significantly higher. This may be explained by steric hindrance between functionalized NP, which are ∼9 nm in diameter plus the flexible GFP-bearing arm. This flexible long arm may, however, favor efficient labeling of adjacent molecules. The fact that the cis-interface disruption significantly decreased the fraction of NP spaced <10 nm, as well as their clustering, strongly suggests that we indeed visualized predicted Ecad arrays stabilized by trans- and cis-interactions in the crystal (Harrison et al., 2011). We frequently observed a few (three to six) particles arranged in straight or broken lines as well as clusters of particles organized in two-dimensional arrays corresponding to the predicted organization of arrays predicted by modeling (Wu et al., 2011). The size of the nanoclusters is below the estimate of cluster size made by Truong Quang et al. (2013) using super-resolution imaging in Drosophila melanogaster embryos, although the comparison is difficult. Indeed, on the one hand, each Ecad molecule within a cluster might not be decorated with a NP and, on the other hand, super-resolutive imaging did not reach nanometric resolution, allowing to address whether Ecad molecules are packed in oligomeric nanoclusters. However, our data fit very well with the estimate of the number of molecules per cluster as well as of the surface of these clusters obtained by Wu et al. (2015) in mammalian cells.
The absence of the cis-interface did not prevent the formation of AJs in epithelial cells. This result recalls modeling data predicting the assembly of membrane-bound ligand–receptor complexes in microdomains (Weikl et al., 2002; Krobath et al., 2011; Bihr et al., 2012; Schmidt et al., 2012). In these models, an initial interaction brings locally the two membranes in close contact, increasing the probability of association of other freely diffusing ligands. This diffusion trap mechanism, similar to the one we proposed earlier for cadherin adhesions (Mège et al., 2006), leads to the buildup of densities of ligand–receptor complexes. Outside of these densities, the spacing of the two membranes superior to the length of the ligand–receptor complex prevents their growth from leading to the formation of discrete regularly spaced clusters. This remoteness has been attributed either to membrane thermal fluctuations or accumulations of membrane-bound glycoproteins. In these models, the increase in cooperatively brought by low energy cis-interactions (Wu et al., 2015) may be negligible. The absence of effect of cis-interface disruption on cadherin adhesion formation further supports this hypothesis. Indeed, cadherin adhesions formed on Ecad-Fc surfaces are found in areas of close contact between the plasma membrane and the substratum, whereas inter-cadherin adhesion areas are characterized by larger membrane-substratum spacing (Lambert et al., 2007). Although we cannot exclude that mutation of the cis-interface slightly destabilizes trans-interactions, the fact that cadherin adhesion and AJ formation are not affected does not advocate for this hypothesis. This is further supported by equilibrium analytical ultracentrifigation data showing that the KD for Ecad ectodomain dimerization is not affected by this mutation (Harrison et al., 2011).
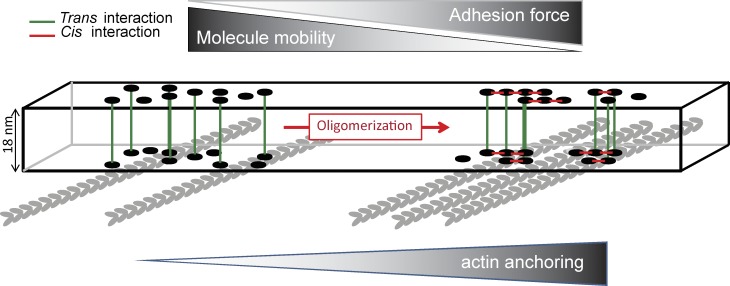
Oligomerization through cis-interactions slightly stabilizes cadherins at cell–cell contacts, in confirmation of previous FRAP experiments performed with tailless cadherins (Harrison et al., 2011). This increase in stability of Ecad was associated with an increase in stability of α-catenin at contact sites, indicating that Ecad ectodomain stability drives the dynamics of its associated partners. The formation of ordered clusters by stabilizing α-catenin may thus favor the association of the clusters to actin filaments. The disruption of the cis-interface significantly increases the retrograde flow of the actin networks in the lamellipodia of cells plated on Ecad-Fc. As reported previously (Plestant et al., 2014), this may result from a weaker coupling of the actin retrograde flow to the adhesion complexes. In addition, the amplitude and the frequency of membrane protrusion are increased in cis-Ecad expressing cells, which could also result from a weaker association to actin. To directly evaluate the anchoring of Ecad clusters to actin we analyzed the mobility of Ecad-Fc–coated beads bound to the surface of transfected cells because the restriction of cadherin mobility in the membrane has been associated with its anchoring to actin (Sako et al., 1998; Lambert et al., 2002). The binding of Ecad-Fc beads was not affected by the oligomeric status of Ecad. In contrast, the bead mobility was significantly higher when the cis-interface was mutated. The displacement of the beads under force was greater for cis-Ecad than for wt Ecad, indicating a weaker mechanical coupling of cellular Ecad to the underlying actin networks. Altogether these data indicate a contribution of oligomerization in the stiffening of the link between Ecad and actin. How can the stabilization of oligomeric clusters have such a strong influence on their anchoring to actin? The slight reduction in α-catenin accumulation may locally alter F-actin dynamics by regulating binding of nucleation and disassembly factors (Hansen et al., 2013). However, an obvious hypothesis is that the organization of Ecad molecules in clusters decreases the probability of rupturing their link to actin by a cooperative effect. Cadherin–catenin complexes within a nonorganized, fluid cluster would behave independently, preventing their cooperative binding to actin (Fig. 9).
Figure 9.
Schematics of the cis-interface–dependent oligomerization of cadherin ectodomain and its effect on F-actin anchoring. The slight stabilization of cell–cell contacts brought by cadherin oligomerization in oligomeric clusters has a strong influence on the anchoring of these clusters to the actomyosin network.
The mechanical stabilization controlled by cadherin oligomerization had strong functional incidence on collective cell behavior. The disruption of the cis-interface increased by >70% of the speed of monolayer expansion. It was associated with a decrease of the order parameter. In other words, the monolayer composed of cis-Ecad cells behaves as a more fluid system. These changes in the fluidity of the monolayer can be directly attributed to a destabilization of the cell–cell contacts facilitating cell–cell contact reshaping and cell partner exchange. Thus, disruption of the cis-interface, albeit having a mild effect on junction formation, drastically alters cell cohesion. In conclusion, we show that cis-interactions stiffen Ecad molecule anchoring to the actin cytoskeleton, allowing cells to acquire more stable contacts and behave more collectively. Thus our results provide direct evidence that cadherin oligomerization indeed supplies the necessary strength to maintain tissue cohesion. Whether this pathway is modulated in vivo during collective cell migration, cell intercalation, wound healing, or cancer cell dissemination by factors such as cadherin glycosylation, cadherin and catenin phosphorylation, or other posttranslational modifications remains to be investigated.
Materials and methods
Cell culture and transient cell transfections
A431D Ecad-deficient epidermoid carcinoma cell line (Lewis et al., 1997; Hong et al., 2010) was grown in DMEM supplemented with 10% FBS, 2 mM l-glutamine, 100 IU of penicillin, and 100 µg/ml streptomycin at 37°C in 5% CO2. The plasmids encoding human wt Ecad-GFP (wt Ecad) and Ecad-V81D-L175D-GFP mutant (cis-Ecad) under the control of the CMV promotor, in the pRC-CMV vector, were derived from the corresponding Ecad constructs fused to Dendra (Harrison et al., 2011) by replacing exactly the Dendra coding sequence with the EGFP coding sequence in the C-terminal of the Ecad coding sequence. Cells were transfected with wt Ecad, cis-Ecad, CAAX-GFP, α-E-catenin-mCherry, and LifeAct-Ruby thanks to the Amaxa Cell Line Nucleofector Kit T (program X-001; Lonza), resulting in >80% transfection efficiency. Assays were performed 24–48 h after transfection. Cells were always used at P < 20.
Protein extraction and coimmunoprecipitation
Proteins were extracted from 5–10 × 106 transfected cells. Cells were rinsed in ice-cold PBS, detached with a cell scraper in cold PBS, and centrifuge at 1,000 rpm for 10 min at 4°C. Whole cell extracts were obtained by lysing cells into cold RIPA buffer (10 mM sodium phosphate buffer, pH 7.8, 60 mM NaCl, 1% Triton X-100, 10% glycerol, 25 mM β-glycerol-phosphate, 50 mM sodium fluoride, 2 mM sodium pyrophosphate, 1 mM orthovanadate, and protease inhibitor cocktail [Complete; Roche]). Lysates were agitated for 15 min at 4°C and cleared by centrifugation at 13,000 rpm for 10 min at 4°C, and protein concentration was estimated by micro-BCA assay (Thermo Fisher Scientific). GFP-tagged proteins were then coimmunoprecipitated using GFP-Trap according to the instructions of the manufacturer (ChromoTek). Protein samples (input and bound) were then subjected to SDS-PAGE and transferred on nitrocellulose membranes at 4°C. Membranes were then blocked with 5% nonfat milk and incubated with the corresponding primary antibody and then with IRDye-coupled secondary antibody (Rockland) against rabbit or mouse immunoglobulins, which were detected with Odyssey Imaging System (LI-COR Biosciences).
Immunocytochemistry
Cells were fixed for 12 min at room temperature using 4% paraformaldehyde in PBS, and then rinsed with PBS and permeabilized for 45 min in PBS supplemented with 1.5% BSA and 0.1% Triton X-100. Cells were then incubated for 1 h with mouse anti–α-catenin (BD) at 1:400 or rabbit anti–β-catenin (Sigma-Aldrich) at 1:400 dilution in PBS-BSA, rinsed, and incubated 1 h with anti–mouse or anti–rabbit Cy3–conjugated antibodies (Jackson ImmunoResearch Laboratories, Inc.) at 1:500 dilution. Preparations were mounted in Mowiol, 90% glycerol, and PBS. Images were taken with a microscope (DM6000; Leica) equipped with a 63× oil objective and Micromax charge coupled device (CCD) camera (Roper Scientific).
Cell membrane fluorescence analysis
Transfected cells were trypsinized, washed, fixed in 3.5% formaldehyde for 15 min, washed again, and imaged under flow using ImageStream X (Amnis) set with the 488-nm laser and 480–560 filter. Data were analyzed using the IDEAS software (Amnis). Regions corresponding to the cell membrane were extracted from bright field images. In brief, two masks were created by eroding and expanding the object by 4 pixels, respectively. The subtraction of the two masks corresponding to the cell membrane region was then applied on the fluorescence image to extract the cell membrane fluorescent intensity. Data acquisition was performed for 300–1,200 cells for each condition and repeated three times.
Ecad-Fc and fibronectin coating procedure
Silanized glass coverslips or electron microscopy Formvar/carbon-coated gold grids (Oxford Instruments SAS) were coated with a human Ecad-human Fc chimera (R&D Systems) or fibronectin (EMD Millipore) as reported previously (Gavard et al., 2004a). In brief, 5 µg of anti–human IgG antibodies (Jackson ImmunoResearch Laboratories, Inc.) in 130 µl Ca2+ Mg2+ PBS were left to adsorb overnight at 4°C. The surfaces were washed three times with PBS, and then 10 µg of Ecad-Fc chimera proteins in PBS were allowed to bind for 2–3 h at room temperature. After three washes, coverslips were blocked for 45 min with PBS and 2.5% BSA.
Preparation of mix-capped gold NPs conjugated to GFP-NPs
Gold NPs used in this study were obtained from British Biocell International Ltd (5-nm diameter). PEGylated alkanethiol, HS-EC11-EG4 (HS-PEG), was purchased from ProChimia Surfaces and the CVVVT-ol peptidol (T-ol is for threoninol) is from Peptide and Protein Research. Mix-capped gold NPs (HS-PEG/CVVVT-ol, ratio 30:70) bearing only one Ni-trisNTA function per NP were prepared as described previously (Lata et al., 2005; Tinazli et al., 2005; Lévy et al., 2006; Duchesne et al., 2008). In brief, Mix-Matrix ligand solution at 2 mM final concentration was prepared by mixing 70 vol of CVVVT-ol at 2 mM with 30 vol of HS-PEG at 2 mM. A controlled molar ratio of 0.01% of HS-C16-EG3-trisNTA functional ligand (2-mM initial concentration) was then added to the Mix-Matrix solution. Capped NPs were prepared by adding 9 vol of colloidal gold solution to 1 vol of the previous ligand solution in a final buffer of PBS supplemented with 0.005% Tween-20 (PBST). Note that the ratio of the trisNTA functional ligand used (0.01% here) has been experimentally calculated to obtain Mix-Matrix–capped NPs bearing no more than one trisNTA group per NP (∼10% of NPs with one trisNTA function and ∼90% with none) and must be adjusted for each new batch of NPs and/or of ligands (matrix or functional). After an overnight incubation on a rotating wheel at room temperature, excess ligands were removed by G25 chromatography using water supplemented with 150 mM NaCl and 0.005% Tween-20 (HNT) as a mobile phase. Then, nickel loading was performed by adding NiCl2 at 250 mM final concentration to the NP solution. After incubation for 1 h on a wheel at room temperature, excess NiCl2 was removed by G25 chromatography using HNT as a mobile phase. Resulting Ni-trisNTA-NPs were purified by affinity chromatography (Histidine-resin). After elution with PBST supplemented with 200 mM imidazole, excess imidazole was removed by G25 chromatography using PBST as a mobile phase. When needed, capped NPs were concentrated by centrifugation at 60,000 g for 30 min. Resulting Ni-trisNTA-NPs were then coupled with polyhistidine-tagged camel anti–GFP-NPs (nanobody GFP-Trap; ChromoTek) as described previously (Duchesne et al., 2012) for polyhistidine-tagged FGF2 protein. In brief, nanobody GFP-Trap (6 µM final concentration) was mixed with purified Ni-trisNTA-NP (200 nM final concentration) in 10 µl PBST (vol/vol). The reaction was left 3 h at room temperature and PBST was added to a final volume of 200 µl. Centrifugation was performed for 90 min at 17,000 g at 4°C, and the supernatant, corresponding to free soluble anti-GFP-NP (uncoupled), was removed. The pellet was resuspended in 200 µl PBST and centrifuged again; a total of five cycles of centrifugation were performed. At the end, the pellet, which corresponds to the purified anti-GFP-NP conjugate (stoichiometry 1:1), was resuspended in PBS at a final concentration of 10 nM. The final concentration of conjugated NPs was calculated using ε520nm of gold NPs given by the manufacturer.
Conjugated NP size
The actual measured diameter of the gold NPs was 6.9 ± 1.8 nm (n = 2,667 NPs from three independent images). They were surrounded by self-assembling monolayers of ∼2.5 nm (Harder et al., 1998; Duchesne et al., 2008), which give an inferred diameter of the functionalized NP of ∼9 nm. The length of the flexible Ni-trisNTA alkyl-OEG-thiol ligand (4.3 nm; Tinazli et al., 2005), of the nanobody itself, and of the GFP of both is in the range of 3 to 4 nm.
TEM experiments
For visualization of individual cadherin molecules, wt Ecad-GFP or cis-Ecad-GFP expressing cells were mechanically detached from the culture flask in the presence of PBS, 3.5 mM EDTA, and 2% BSA on ice and allowed to adhere for 2 h on Ecad-Fc– or fibronectin-coated electron microscopy grids, at 37°C in the absence of serum. After three washes with DMEM to remove the nonadherent cells, plasma membrane sheets on the electron microscopy grids were prepared (“rip-off” procedure) as described previously (Prior et al., 2003; Hancock and Prior, 2005) with some modifications. In brief, cells on grids were pressed onto a clean glass coverslip. The coverslip was turned over and 200 µl PBS was added quickly around the grids to separate them from the coverslip and to generate plasma membrane sheets on the grids (inner leaflet face up). Samples were then fixed with a solution of 0.1% glutaraldehyde (vol/vol) and 4% formaldehyde (wt/vol) for 10 min. The fixative was then quenched with three washes in 100 mM glycine. After three bathes in PBS, nonspecific sites were blocked for 10 min with PBS supplemented with 0.25% BSA. Grids were then incubated for 30 min with 3 nM anti-GFP-NP (or control uncoupled Ni-trisNTA-NP) in PBS and 0.25% BSA. After extensive washes with PBS and then deionized water, grids were treated with a mixture of 0.3% uranyl acetate (wt/vol) and 1.8% methylcellulose (wt/vol) for 10 min on ice. Grids were then individually picked up with homemade 5-mm-thick iron wire loops and left to dry overnight before storage. Preparations were digitally imaged using an 80-kV transmission electron microscope (CM100; Philips) equipped with an Orius CCD Camera (Gatan) or a 200-kV (Tecnai G2 T20 Sphera; FEI) transmission electron microscope equipped with a LaB6 electron source and a USC4000 CCD camera (Gatan). For the analysis, 3-µm2 images acquired with the CM100 were cropped down to 0.55-µm2 area to fit with the scale of the ones acquired with the Tecnai G2. Four and three different grids were visualized for wt and cis conditions, respectively. All 0.55-µm2 images (742.7 × 742.7 nm) were processed using Fiji software (ImageJ, National Institutes of Health). In brief, digital pictures were converted to binary images and filtered to remove any residual noise, and then x,y coordinates of the NPs were calculated for each picture. To avoid bias that would be caused by a small number of pictures having a very high density of gold NP labeling, only images with 2 to 100 gold NPs per pictures (4–180 NP/µm2) were used for further analysis. For oligomer counting, given a mean diameter for the functionalized NPs (∼9 nm) and the length of the flexible Ni-TrisNTA ligand (∼4.3 nm; see previous section), a maximal distance of 15 nm between NP centers was fixed to distinguish cadherin ectodomains in interaction from non-interacting monomers in cis. Calculation of the distance between each NP and its nearest neighbor (center to center) was performed using the NND (Nearest Neighbor Distance) imageJ plugin (from Y. Mao, Mississippi State University, Starville, MS). Cluster analysis was performed using the Univariate 725 macro (I. Prior, University of Liverpool, Liverpool, UK) as described in Hancock and Prior (2005). For such analysis, only fairly homogeneous images without vesicular profiles or large unlabeled areas were kept (n = 50 and 102 for wt and cis-Ecad expressing cells, respectively). In brief, K-function identifies systematic deviations of the NPs pattern from complete spatial randomness. The mean K-function is plotted as a linear transformation L(r) − r. To interpret the statistical significance indicating clustering, a 99% confidence interval for L(r) − r is generated using Monte Carlo simulations. Plots were generated for each individual image and L(r) − r values were then standardized on the confidence interval for each image to allow comparison and averaging. Averaging was done for all images and specifically for images presenting a significant clustering between 0- to 300-nm radius. Values above 1 for the standardized L(r) − r function indicate significant clustering (99% confidence interval) at the radius r and no deviation (0 < L(r) − r < 1) indicates a random pattern.
Graphs and statistical analyses were performed using OriginPro 8.6 software. Kolmogorov-Smirnov nonparametric test was used to compare distribution and χ2 test was performed to compare proportions.
For analysis of intercellular junctions, transfected cells were deposited on a 12-well format cell culture insert at high density (pore size of 0.4 µm; BD). When the monolayer was confluent, cells were fixed with 3% glutaraldehyde in phosphate buffer for 1 h at room temperature. Samples were kept at 4°C in 0.1 M phosphate buffer, pH 7.4, until further treatment and embedding in epon resin was performed. Thin section chromatography was performed and samples were stained with uranyl acetate and observed using a transmission electron microscope (1011; JEOL) equipped with an Orius CCD camera.
FRAP
FRAP was measured at 37°C on cells coelectroporated with wt Ecad-GFP or cis-Ecad-GFP and αE-catenin-mCherry using a confocal microscope (TCS SP5; Leica) equipped with a 40× water immersion objective. After five prebleach scans (0.347 s), a rectangular region of interest (3.5 × 2.9 µm) was bleached and fluorescence recovery was acquired every 0.347 s (20 scans), then every 2 s (20 scans), and finally every 10 s (20 scans). The normalized recovery of fluorescence was expressed as a ratio of prebleach fluorescence rate after correction for photobleaching, as reported previously (Lambert et al., 2007). Fluorescence recovery in function of time were best fitted with a one-term exponential equation, allowing to extract a plateau value representing the fraction of diffusion-limited molecules (mobile fraction) and a recovery t1/2 proportional to the apparent diffusion coefficient of diffusion-limited molecules (Thoumine et al., 2006). The mobile fraction and the t1/2 were determined by fitting the normalized recovery curves using one-phase decay nonlinear regression function of the Prism 5.01 software (GraphPad Software).
Collective cell migration assay
A431D cells expressing wt Ecad or cis-Ecad were high density plated in 3.5-cm Petri dishes where a polydimethylsiloxane (PDMS) block was previously deposited to impose cells to grow on a restricted area of the dish. When cells reached confluence, the PDMS block was removed. Images were then acquired every 5 min during 24 h under a controlled temperature and CO2 environment (5% CO2 at 37°C; 10× objective; BioStation; Nikon). The surface occupied by the monolayer determined thanks to ImageJ was plotted as a function of time. Manual tracking of individual cells at the front or rear (at least four rows of cells away from the front) was performed with the MTrackJ plugin during the first 8 h. Individual trajectories were positioned on an orthonormal axis with the coordinates of the cell at t0 = (0, 0). The MSD was then extracted for each condition and plotted versus time. The direction persistence was calculated as the ratio of the cumulative distance over the Euclidian distance between the position of the cell at time 0 and its position at time t.
Actin dynamics and lamellipodial activity measurement
Cells coexpressing LifeAct-Ruby and wt Ecad-GFP or cis-Ecad-GFP were mechanically detached as described previously (Plestant et al., 2014) and plated in live cell imaging buffer (10 mM Hepes, pH 7.4, 128 mM NaCl, 6 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 5.5 mM glucose, and 0.2% BSA) at low cell density (<5 × 104 cells/cm2) on Ecad-Fc–coated glass-bottom dishes for 2 h. Cells were then imaged every 500 ms for 3 min at 63× with a time-lapse confocal video microscope equipped with a Nipkov disk (spinning disk). Kymographs were made by generating time-lapse montages of a single line perpendicular to the cell edge for each frame of the video (ImageJ) along three lines normal to the free edge of each analyzed cell. Three flow rates were calculated for each kymograph (15 cells analyzed for each condition).
Preparation of the Ecad-coated beads
2.8 µm of magnetic protein A–coated beads (Dynabeads; Invitrogen) were coated with Ecad-hFc. In brief, 10 µl of the blurry solution was washed three times and resuspended in 200 µl of 0.1 M borate buffer, pH 8.0, before 2 × 30-s sonication. Then, 50 µl of goat anti–human IgG Fc fragment (2.4 mg/ml; Jackson ImmunoResearch Laboratories, Inc.) was added and left to incubate overnight on a wheel at room temperature. Beads are then washed three times and resuspended in 200 µl PBS (Life technologies) before 2 × 30-s sonication. Then 5 µl of recombinant Ecad-Fc were added and left to incubate for 3 h on a rotating wheel at room temperature. Finally, beads were washed and resuspended in 1 ml PBS supplemented with 1% BSA. 50 µl of this Ecad-coated bead solution was added to cells grown on a 22 × 22-mm glass coverslip placed on a 3.5-cm Petri dish for 1 h. After extensive washes to remove unbound beads, the medium was changed for phenol red–free DMEM supplemented with 20 mM Hepes, pH 7.4.
For bead binding assays, Ecad-hFc– or hFc-coated beads were deposited on nontransfected and wt Ecad– or cis-Ecad–transfected cells, left to adhere for 1 h, and gently washed before fixation. Images were taken with a DM6000 microscope equipped with a 10× objective and a micromax CCD camera. The number of bound beads per squared millimeter was then manually scored.
Magnetic tweezers assay
The forces were locally applied on bead-bearing cells with magnetic tweezers made of an electromagnet and the superparamagnetic microbeads mentioned in Preparation of the Ecad-coated beads. The electromagnet was 816 turns of 0.5-mm copper wire coil surrounding a soft iron core 5 mm in diameter with a 30° cone-shaped tip. It was mounted on a micromanipulator (InjectMan NI2; Eppendorf) at a 45° vertical angle, and the tip initially aligned at 700 µm from the center of the observation zone. The current was provided by a home-made voltage-controlled current and a function generator (TG1010; TT Instruments). This function generator was directly controlled from the computer through a control card (USB1208HS; Measurement Computing). The samples were mounted on a microscope (DMIRB; Leica) equipped with a CCD camera (Coolsnap HQ2; Roper Scientific) through a 100× oil objective. Both the camera and the current in the coil were controlled by the µManager software (version 1.4.8). 2.8 µm of Ecad-Fc–coated magnetic beads were preincubated for 1 h on wt or cis-Ecad cells, and then the unbound beads were washed away. Applications of current ranging from 0.5 to 1.2 A drew the beads toward the tip. The force exerted by the electromagnet was calibrated by measuring the velocity of a bead moving through a viscous fluid (PDMS). Six steps of forces were applied, each step consisted of the following: at t = 0, current in the coil was set to 1.2 A and camera received signal to start acquiring images in the burst mode (frequency of ∼13 frames/s) for 170 frames; the current was turned down and the camera was set to an acquiring rate of 2 frames/s for 114 s; the camera was then set to a 1-frame/s acquisition rate for an additional 125 s. Tracking of the bead position was done with Icy (Icy v1.4.3.5; Quantitative Image Analysis Unit, Institut Pasteur) using the Active Contour plugin.
PIV
PIV is an image correlation–based method usually used to obtain instantaneous velocity field measurements from the local displacements and heavily used in hydrodynamics. Images are divided into multiple interrogation subwindows. Each interrogation subwindow should contain sufficient numbers of tracers to enable comparison between the current time frame and the subsequent time frame. Cross-correlation techniques are then performed to compute the displacement vectors at each subwindow by finding their best match at the successive time frame. PIV analysis of monolayer movement (Petitjean et al., 2010) was performed as described previously (Vedula et al., 2012) using MatPIV v. 1.6.1 package and implemented in Matlab (MathWorks). The analysis was done with 32 × 32-pixel (19 × 19-µm) interrogation windows with an overlap of 50% using the same size of initial migrating front (300 µm). Order parameter and correlation length were calculated using the formula previously described (Doxzen et al., 2013; Vedula et al., 2014).
Statistical analysis and curve fitting and image processing
Statistical analysis and curves fitting were performed with Prism 5.0 software. Differences were considered significant for p-values ≤0.05. Image processing was done in ImageJ (or Matlab when indicated), and then with Photoshop and Illustrator (Adobe).
Online supplemental material
Fig. S1 shows the recruitment of β-catenin at cell–cell contacts and binding of Ecad-Fc beads independent of Ecad cis-oligomerization. Fig. S2 shows the nanometric organization of wt and cis-Ecad-GFP at the cell membrane of A431D-transfected cells. Fig. S3 shows the quantitative analysis of wt and cis-Ecad-GFP cell layer expansion. Fig. S4 shows the migration of single wt and cis-Ecad-GFP–transfected cells. Fig. S5 shows the division rate of wt and cis-Ecad-GFP–transfected cells. Videos 1 and 2 show actin retrograde flow in wt Ecad-GFP expressing cell spread on Ecad-Fc (Video 1) and in cis-Ecad-GFP expressing cell spread on Ecad-Fc (Video 2). Videos 3 and 4 show Ecad-Fc magnetic bead displacement under force in wt Ecad-GFP (Video 3) and in cis-Ecad-GFP expressing cells (Video 4). Videos 5 and 6 show collective cell migration of wt Ecad-GFP (Video 5) and of cis-Ecad-GFP expressing cells (Video 6). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201410111/DC1.
Supplementary Material
Acknowledgments
We thank N. Boggetto for her help with flow imaging and D. Chrétien and A. Schmitt for help with electron microscopy. We thank Ian Prior for providing the Univariate 725 Excel macro.
This work was supported by grants from Centre National de la Recherche Scientifique, Fondation ARC (R.M. Mège), Human Frontier Science Program grant RPG0040/2012 (B. Ladoux and R.M. Mège), Agence Nationale de la Recherche (ANR 2010 Blan1515 [B. Ladoux and R.M. Mège] and Nanotechnology Program [B. Ladoux]), and European Research Council (ERC) under the European Union’s Seventh Framework Program (FP7/2007-2013)/ERC grant agreement no. 617233 (B. Ladoux). Financial supports were from Mechanobiology Institute and Institut Universitaire de France to B. Ladoux. S. Troyanovsky’s work was supported by grant AR44016 from the National Institutes of Health. G. Peyret, P.-O. Strale, and L. Duchesne were supported by fellowships from the University Paris-Diderot, Agence Nationale de la Recherche, and Fondation ARC, respectively. We acknowledge the Institut Jacques Monod ImagoSeine, Microscopy Rennes imaging center, and Institut Cochin Electron Microscopy facilities.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AJ
- adherens junction
- MSD
- mean square displacement
- NP
- nanoparticle
- PDMS
- polydimethylsiloxane
- PIV
- particle image velocimetry
- TEM
- transmission electron microscopy
- wt
- wild type
References
- Adams C.L., Chen Y.T., Smith S.J., and Nelson W.J.. 1998. Mechanisms of epithelial cell–cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin–green fluorescent protein. J. Cell Biol. 142:1105–1119. 10.1083/jcb.142.4.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Amoudi A., Díez D.C., Betts M.J., and Frangakis A.S.. 2007. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 450:832–837. 10.1038/nature05994 [DOI] [PubMed] [Google Scholar]
- Baumgartner W., Hinterdorfer P., Ness W., Raab A., Vestweber D., Schindler H., and Drenckhahn D.. 2000. Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 97:4005–4010. 10.1073/pnas.070052697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihr T., Seifert U., and Smith A.-S.. 2012. Nucleation of ligand-receptor domains in membrane adhesion. Phys. Rev. Lett. 109:258101 10.1103/PhysRevLett.109.258101 [DOI] [PubMed] [Google Scholar]
- Boggon T.J., Murray J., Chappuis-Flament S., Wong E., Gumbiner B.M., and Shapiro L.. 2002. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 296:1308–1313. 10.1126/science.1071559 [DOI] [PubMed] [Google Scholar]
- Cavey M., Rauzi M., Lenne P.F., and Lecuit T.. 2008. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 453:751–756. 10.1038/nature06953 [DOI] [PubMed] [Google Scholar]
- Doxzen K., Vedula S.R.K., Leong M.C., Hirata H., Gov N.S., Kabla A.J., Ladoux B., and Lim C.T.. 2013. Guidance of collective cell migration by substrate geometry. Integr. Biol. (Camb.). 5:1026–1035. 10.1039/c3ib40054a [DOI] [PubMed] [Google Scholar]
- Duchesne L., Gentili D., Comes-Franchini M., and Fernig D.G.. 2008. Robust ligand shells for biological applications of gold nanoparticles. Langmuir. 24:13572–13580. 10.1021/la802876u [DOI] [PubMed] [Google Scholar]
- Duchesne L., Octeau V., Bearon R.N., Beckett A., Prior I.A., Lounis B., and Fernig D.G.. 2012. Transport of fibroblast growth factor 2 in the pericellular matrix is controlled by the spatial distribution of its binding sites in heparan sulfate. PLoS Biol. 10:e1001361 10.1371/journal.pbio.1001361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavard J., Lambert M., Grosheva I., Marthiens V., Irinopoulou T., Riou J.F., Bershadsky A., and Mège R.M.. 2004a Lamellipodium extension and cadherin adhesion: two cell responses to cadherin activation relying on distinct signalling pathways. J. Cell Sci. 117:257–270. 10.1242/jcs.00857 [DOI] [PubMed] [Google Scholar]
- Gavard J., Marthiens V., Monnet C., Lambert M., and Mège R.M.. 2004b N-cadherin activation substitutes for the cell contact control in cell cycle arrest and myogenic differentiation: involvement of p120 and β-catenin. J. Biol. Chem. 279:36795–36802. 10.1074/jbc.M401705200 [DOI] [PubMed] [Google Scholar]
- Giannone G., Mège R.M., and Thoumine O.. 2009. Multi-level molecular clutches in motile cell processes. Trends Cell Biol. 19:475–486. 10.1016/j.tcb.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Goodsell D.S., and Olson A.J.. 2000. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 29:105–153. 10.1146/annurev.biophys.29.1.105 [DOI] [PubMed] [Google Scholar]
- Hancock J.F., and Prior I.A.. 2005. Electron microscopic imaging of Ras signaling domains. Methods. 37:165–172. 10.1016/j.ymeth.2005.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S.D., Kwiatkowski A.V., Ouyang C.Y., Liu H., Pokutta S., Watkins S.C., Volkmann N., Hanein D., Weis W.I., Mullins R.D., and Nelson W.J.. 2013. αE-catenin actin-binding domain alters actin filament conformation and regulates binding of nucleation and disassembly factors. Mol. Biol. Cell. 24:3710–3720. 10.1091/mbc.E13-07-0388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder P., Grunze M., and Dahint R.. 1998. Molecular conformation in oligo(ethylene glycol)-terminated self-assembled monolayers on gold and sylver surfaces determines their ability to resist protein adsorption. J. Phys. Chem. 102:426–436. 10.1021/jp972635z [DOI] [Google Scholar]
- Harrison O.J., Jin X., Hong S., Bahna F., Ahlsen G., Brasch J., Wu Y., Vendome J., Felsovalyi K., Hampton C.M., et al. . 2011. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure. 19:244–256. 10.1016/j.str.2010.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Cowin P., and Stokes D.L.. 2003. Untangling desmosomal knots with electron tomography. Science. 302:109–113. 10.1126/science.1086957 [DOI] [PubMed] [Google Scholar]
- Hong S., Troyanovsky R.B., and Troyanovsky S.M.. 2010. Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc. Natl. Acad. Sci. USA. 107:3528–3533. 10.1073/pnas.0911027107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Troyanovsky R.B., and Troyanovsky S.M.. 2013. Binding to F-actin guides cadherin cluster assembly, stability, and movement. J. Cell Biol. 201:131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulpiau P., Gul I.S., and van Roy F.. 2013. New insights into the evolution of metazoan cadherins and catenins. Prog. Mol. Biol. Transl. Sci. 116:71–94. 10.1016/B978-0-12-394311-8.00004-2 [DOI] [PubMed] [Google Scholar]
- Kollmannsberger P., and Fabry B.. 2007. High-force magnetic tweezers with force feedback for biological applications. Rev. Sci. Instrum. 78:114301 10.1063/1.2804771 [DOI] [PubMed] [Google Scholar]
- Krobath H., Różycki B., Lipowsky R., and Weikl T.R.. 2011. Line tension and stability of domains in cell-adhesion zones mediated by long and short receptor-ligand complexes. PLoS ONE. 6:e23284 10.1371/journal.pone.0023284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert M., Choquet D., and Mège R.M.. 2002. Dynamics of ligand-induced, Rac1-dependent anchoring of cadherins to the actin cytoskeleton. J. Cell Biol. 157:469–479. 10.1083/jcb.200107104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert M., Thoumine O., Brevier J., Choquet D., Riveline D., and Mège R.M.. 2007. Nucleation and growth of cadherin adhesions. Exp. Cell Res. 313:4025–4040. 10.1016/j.yexcr.2007.07.035 [DOI] [PubMed] [Google Scholar]
- Lata S., Reichel A., Brock R., Tampé R., and Piehler J.. 2005. High-affinity adaptors for switchable recognition of histidine-tagged proteins. J. Am. Chem. Soc. 127:10205–10215. 10.1021/ja050690c [DOI] [PubMed] [Google Scholar]
- le Duc Q., Shi Q., Blonk I., Sonnenberg A., Wang N., Leckband D., and de Rooij J.. 2010. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II–dependent manner. J. Cell Biol. 189:1107–1115. 10.1083/jcb.201001149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy R., Wang Z., Duchesne L., Doty R.C., Cooper A.I., Brust M., and Fernig D.G.. 2006. A generic approach to monofunctionalized protein-like gold nanoparticles based on immobilized metal ion affinity chromatography. ChemBioChem. 7:592–594. 10.1002/cbic.200500457 [DOI] [PubMed] [Google Scholar]
- Lewis J.E., Wahl J.K. III, Sass K.M., Jensen P.J., Johnson K.R., and Wheelock M.J.. 1997. Cross-talk between adherens junctions and desmosomes depends on plakoglobin. J. Cell Biol. 136:919–934. 10.1083/jcb.136.4.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mege R.M., Matsuzaki F., Gallin W.J., Goldberg J.I., Cunningham B.A., and Edelman G.M.. 1988. Construction of epithelioid sheets by transfection of mouse sarcoma cells with cDNAs for chicken cell adhesion molecules. Proc. Natl. Acad. Sci. USA. 85:7274–7278. 10.1073/pnas.85.19.7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mège R.M., Gavard J., and Lambert M.. 2006. Regulation of cell–cell junctions by the cytoskeleton. Curr. Opin. Cell Biol. 18:541–548. 10.1016/j.ceb.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Mitchison T., and Kirschner M.. 1988. Cytoskeletal dynamics and nerve growth. Neuron. 1:761–772. 10.1016/0896-6273(88)90124-9 [DOI] [PubMed] [Google Scholar]
- Overduin M., Harvey T.S., Bagby S., Tong K.I., Yau P., Takeichi M., and Ikura M.. 1995. Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science. 267:386–389. 10.1126/science.7824937 [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M., Jamora C., and Fuchs E.. 2003. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 112:535–548. 10.1016/S0092-8674(03)00108-9 [DOI] [PubMed] [Google Scholar]
- Perret E., Leung A., Feracci H., and Evans E.. 2004. Trans-bonded pairs of E-cadherin exhibit a remarkable hierarchy of mechanical strengths. Proc. Natl. Acad. Sci. USA. 101:16472–16477. 10.1073/pnas.0402085101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean L., Reffay M., Grasland-Mongrain E., Poujade M., Ladoux B., Buguin A., and Silberzan P.. 2010. Velocity fields in a collectively migrating epithelium. Biophys. J. 98:1790–1800. 10.1016/j.bpj.2010.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plestant C., Strale P.O., Seddiki R., Nguyen E., Ladoux B., and Mège R.M.. 2014. Adhesive interactions of N-cadherin limit the recruitment of microtubules to cell-cell contacts through organization of actomyosin. J. Cell Sci. 127:1660–1671. 10.1242/jcs.131284 [DOI] [PubMed] [Google Scholar]
- Prior I.A., Muncke C., Parton R.G., and Hancock J.F.. 2003. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160:165–170. 10.1083/jcb.200209091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviola E., and Gilula N.B.. 1975. Intramembrane organization of specialized contacts in the outer plexiform layer of the retina. A freeze-fracture study in monkeys and rabbits. J. Cell Biol. 65:192–222. 10.1083/jcb.65.1.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y., Nagafuchi A., Tsukita S., Takeichi M., and Kusumi A.. 1998. Cytoplasmic regulation of the movement of E-cadherin on the free cell surface as studied by optical tweezers and single particle tracking: corralling and tethering by the membrane skeleton. J. Cell Biol. 140:1227–1240. 10.1083/jcb.140.5.1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Bihr T., Seifert U., and Smith A.-S.. 2012. Coexistence of dilute and densely packed domains of ligand-receptor bonds in membrane adhesion. Europhys. Lett. 99:38003 10.1209/0295-5075/99/38003 [DOI] [Google Scholar]
- Shapiro L., and Weis W.I.. 2009. Structure and biochemistry of cadherins and catenins. Cold Spring Harb. Perspect. Biol. 1:a003053 10.1101/cshperspect.a003053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L., Fannon A.M., Kwong P.D., Thompson A., Lehmann M.S., Grübel G., Legrand J.F., Als-Nielsen J., Colman D.R., and Hendrickson W.A.. 1995. Structural basis of cell-cell adhesion by cadherins. Nature. 374:327–337. 10.1038/374327a0 [DOI] [PubMed] [Google Scholar]
- Takeichi M. 2014. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15:397–410. 10.1038/nrm3802 [DOI] [PubMed] [Google Scholar]
- Tambe D.T., Hardin C.C., Angelini T.E., Rajendran K., Park C.Y., Serra-Picamal X., Zhou E.H., Zaman M.H., Butler J.P., Weitz D.A., et al. . 2011. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 10:469–475. 10.1038/nmat3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W.A., Boscher C., Chu Y.S., Cuvelier D., Martinez-Rico C., Seddiki R., Heysch J., Ladoux B., Thiery J.P., Mege R.M., and Dufour S.. 2013. α-Catenin and vinculin cooperate to promote high E-cadherin-based adhesion strength. J. Biol. Chem. 288:4957–4969. 10.1074/jbc.M112.403774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoumine O., Lambert M., Mège R.M., and Choquet D.. 2006. Regulation of N-cadherin dynamics at neuronal contacts by ligand binding and cytoskeletal coupling. Mol. Biol. Cell. 17:862–875. 10.1091/mbc.E05-04-0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinazli A., Tang J., Valiokas R., Picuric S., Lata S., Piehler J., Liedberg B., and Tampé R.. 2005. High-affinity chelator thiols for switchable and oriented immobilization of histidine-tagged proteins: a generic platform for protein chip technologies. Chemistry. 11:5249–5259. 10.1002/chem.200500154 [DOI] [PubMed] [Google Scholar]
- Troyanovsky R.B., Indra I., Chen C.S., Hong S., and Troyanovsky S.M.. 2015. Cadherin controls nectin recruitment into adherens junctions by remodeling the actin cytoskeleton. J. Cell Sci. 128:140–149. 10.1242/jcs.161588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong Quang B.A., Mani M., Markova O., Lecuit T., and Lenne P.F.. 2013. Principles of E-cadherin supramolecular organization in vivo. Curr. Biol. 23:2197–2207. 10.1016/j.cub.2013.09.015 [DOI] [PubMed] [Google Scholar]
- Vedula S.R.K., Leong M.C., Lai T.L., Hersen P., Kabla A.J., Lim C.T., and Ladoux B.. 2012. Emerging modes of collective cell migration induced by geometrical constraints. Proc. Natl. Acad. Sci. USA. 109:12974–12979. 10.1073/pnas.1119313109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedula S.R.K., Hirata H., Nai M.H., Brugués A., Toyama Y., Trepat X., Lim C.T., and Ladoux B.. 2014. Epithelial bridges maintain tissue integrity during collective cell migration. Nat. Mater. 13:87–96. 10.1038/nmat3814 [DOI] [PubMed] [Google Scholar]
- Weikl T.R., Andelman D., Komura S., and Lipowsky R.. 2002. Adhesion of membranes with competing specific and generic interactions. Eur Phys J E Soft Matter. 8:59–66. 10.1140/epje/i2002-10008-2 [DOI] [PubMed] [Google Scholar]
- Wu Y., Vendome J., Shapiro L., Ben-Shaul A., and Honig B.. 2011. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 475:510–513. 10.1038/nature10183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Honig B., and Ben-Shaul A.. 2013. Theory and simulations of adhesion receptor dimerization on membrane surfaces. Biophys. J. 104:1221–1229. 10.1016/j.bpj.2013.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Kanchanawong P., and Zaidel-Bar R.. 2015. Actin-delimited adhesion-independent clustering of E-cadherin forms the nanoscale building blocks of adherens junctions. Dev. Cell. 32:139–154. 10.1016/j.devcel.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Yonemura S., Wada Y., Watanabe T., Nagafuchi A., and Shibata M.. 2010. α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12:533–542. 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.