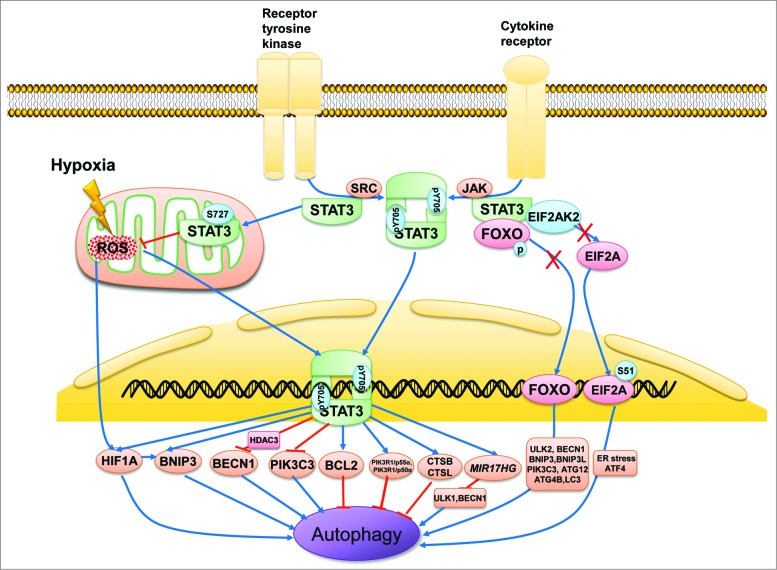
Figure 2.
Subcellular localization of STAT3 in the regulation of autophagy. STAT3 monomers in the cytosol are phosphorylated by Src or JAK kinase on Tyr705 and subsequently form STAT3 dimers, which are then shuttled into the cell nucleus and bind with specific DNA elements to transcriptionally activate or suppress target genes such as BCL2, BECN1, PIK3C3, CTSB, CTSL, PIK3R1/p55α, PIK3R1/p50α, and MIR17HG as well as HIF1A and BNIP3, which either inhibit or stimulate autophagy depending on the different cellular context or stimulus. In contrast, unphosphorylated cytoplasmic STAT3 sequesters EIF2AK2, FOXO1, and FOXO3. EIF2AK2 promotes autophagy by phosphorylating EIF2A, an endoplasmic reticulum stress-related protein that upregulates ATF4. FOXO1 and FOXO3 also positively modulate autophagy by transcriptionally activating a series of autophagy-related genes such as ULK2, BECN1, BNIP3, BNIP3L, PIK3C3, ATG12, ATG4B, and MAP1LC3A. STAT3 monomers also translocate to the mitochondria and interact with complexes I and II of the ETC to repress ROS production. ROS induces autophagy by activating HIF1A, possibly by modulating the transcriptional ability of STAT3.