Figure 1.
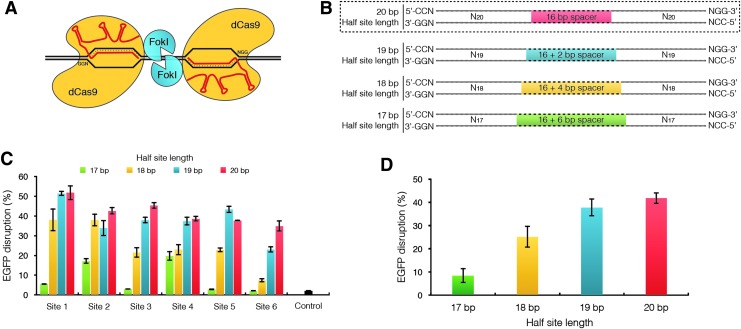
Optimization of dimeric tru-RFN architecture. (A) Schematic of dimeric RNA-guided FokI-dCas9 nucleases (RFNs) binding to DNA. Two FokI-dCas9 proteins are targeted to adjacent PAM-out DNA half-site sequences by two gRNAs enabling FokI dimerization and cleavage of the intervening spacer sequence. (B) Standard RFN and tru-RFN-binding sites. The overall length of the dimeric target site is 62 bp with the 5′-NGG-3′ PAMs facing outward. The spacer length is dependent on the gRNA half-site length: 20, 19, 18, and 17 bp half-sites correspond to 16, 18, 20, and 22 bp spacers, respectively. (C) Mutagenic activity of standard RFNs or tru-RFNs with 17–20 bp half-site lengths. All RFNs are targeted to the same six target sites within the EGFP gene (Supplementary Table S1). Control is U2OS.EGFP nucleofected with ptdTomato-N1 plasmid. EGFP disruption in U2OS.EGFP cells was measured by flow cytometry. Error bars represent s.e.m., n=3. (D) Mean EGFP disruption values of (C) grouped according by gRNA half-site length. Error bars represent s.e.m., n=6. gRNAs, guide RNAs; RFN, RNA-guided FokI-dCas9 nucleases.