Figure 4. Identification of direct HIF1α targets in E12.5 fCMs.
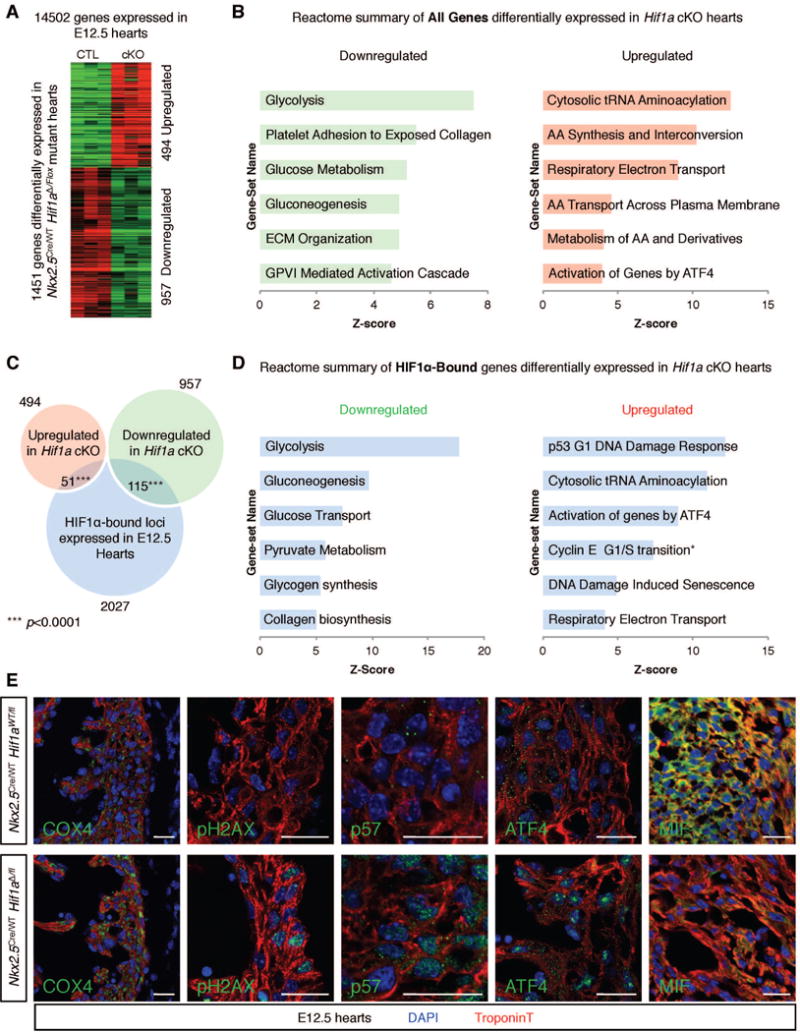
A) Comparison of control and Hif1α cKO E12.5 cardiac transcriptomes (n=3 for each genotype) unveiled a total of 1451 genes modulated in mutant hearts. B) REACTOME database functional clustering of genes down- and up-regulated in cKOs, highlighting cellular processes most significantly affected in mutant hearts. C) Overlay of RNA-seq and ChIP-seq results revealed 166 likely direct targets of HIF1α. D) REACTOME functional clustering of these genes allowed for identification of cellular functions directly regulated by HIF1α. *category containing the G1/S transition inhibitor p21. E) Immunostaining validation of RNA-seq results in the left ventricular free wall or IVS (MIF) of E12.5 hearts. Bars represent 20 μm. AA=amino acid; ECM=extracellular matrix; GPVI=glycoprotein VI. See also Figure S3 and Tables S2, S3 and S4.
