Figure 5. Intersections between HIF1α, ATF4 and p53.
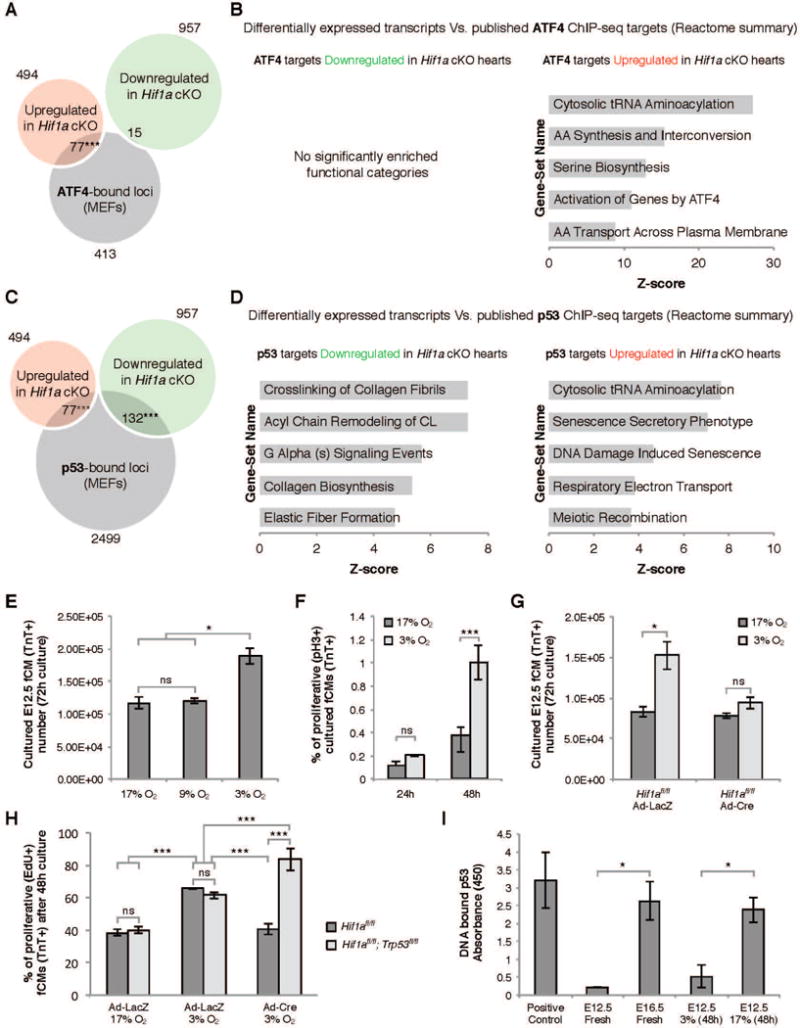
A) Overlap between genes modulated in Hif1a cKO hearts and ATF4 ChIP-seq data revealed a significant enrichment in ATF4 targets amongst upregulated genes. B) REACTOME functional clustering of ATF4 targets modulated in Hif1a cKO hearts. C) Overlap between genes modulated in Hif1a cKO hearts and p53 ChIP-seq data revealed a significant enrichment in p53 targets amongst both down and upregulated genes. D) REACTOME functional clustering of p53 targets modulated in Hif1a cKO hearts. E) Ex vivo culture of primary E12.5 ventricular cells in different oxygen concentrations revealed that hypoxia (3% O2) promoted an increase in fCM number. This effect was associated with increased proliferation of TroponinT-positive fCMs (F) and was dependent on HIF1α (G). H) Blunted proliferation resulting from absence of HIF1α was further demonstrated by quantification of EdU-positive fCMs and could be rescued by the simultaneous ablation of HIF1α and p53. I) Elisa quantification of p53-DNA binding revealed that during cardiogenesis p53 and HIF1α activity inversely correlated. E–I: data represented as mean ± SD; *P<0.05; ***P<0.05. See also Figure S4 and Table S5.
