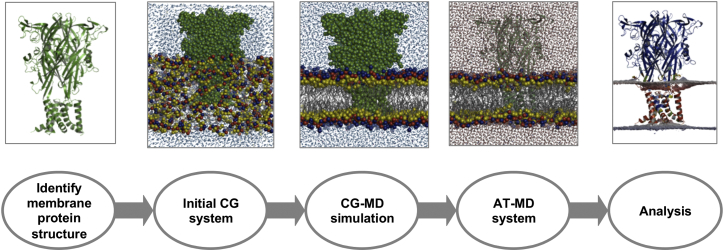
Figure 1.
The MemProtMD Pipeline for Inserting Membrane Proteins into Bilayers
The first step is to detect the protein structures from the PDB, here shown for the P2X4 receptor (PDB: 4DW0, trimeric biological assembly as annotated in the PDB). The second is to set up a lipid, water, and protein simulation system. CG simulations are then run (1 μs duration) to assemble and equilibrate a bilayer around each membrane protein structure. The CG simulation system is then converted to atomic resolution, allowing detailed analysis of lipid bilayer/protein interactions. See also Figure S1.