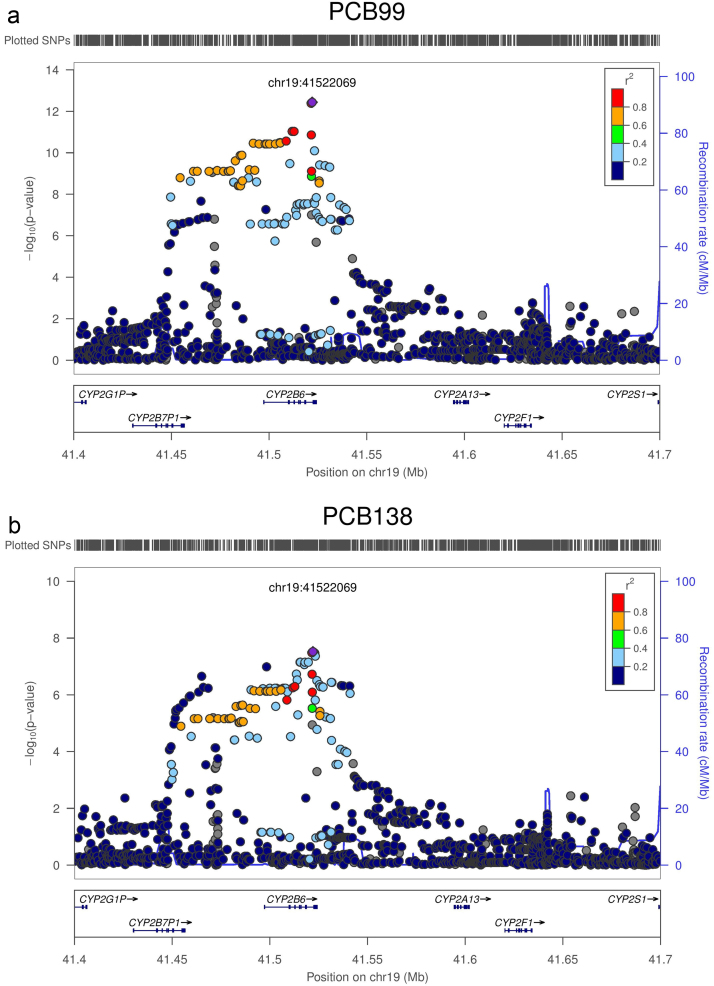
Fig. 2.
(a) Regional Association plot of PCB 99. Each point represents a SNP plotted with their p-value (on a −log10 scale) as a function of genomic position (NCBI Build 37). In each panel, the lead SNP is represented by the purple diamond. The colour coding of all other SNPs (circles) indicates LD with the lead SNP (estimated by CEU r2 from the 1000 Genomes Project March 2012 release): red r2≥0.8; gold 0.6≤r2<0.8; green 0.4≤r2<0.6; cyan 0.2≤r2<0.4; blue r2<0.2; grey r2 unknown. Recombination rates are estimated from the International HapMap Project and gene annotations are taken from the University of California Santa Cruz genome browser. (b) Regional Association plot of PCB 138. Each point represents a SNP plotted with their p-value (on a −log10 scale) as a function of genomic position (NCBI Build 37). In each panel, the lead SNP is represented by the purple diamond. The colour coding of all other SNPs (circles) indicates LD with the lead SNP (estimated by CEU r2 from the 1000 Genomes Project March 2012 release): red r2≥0.8; gold 0.6≤r2<0.8; green 0.4≤r2<0.6; cyan 0.2≤r2<0.4; blue r2<0.2; grey r2 unknown. Recombination rates are estimated from the International HapMap Project and gene annotations are taken from the University of California Santa Cruz genome browser. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)