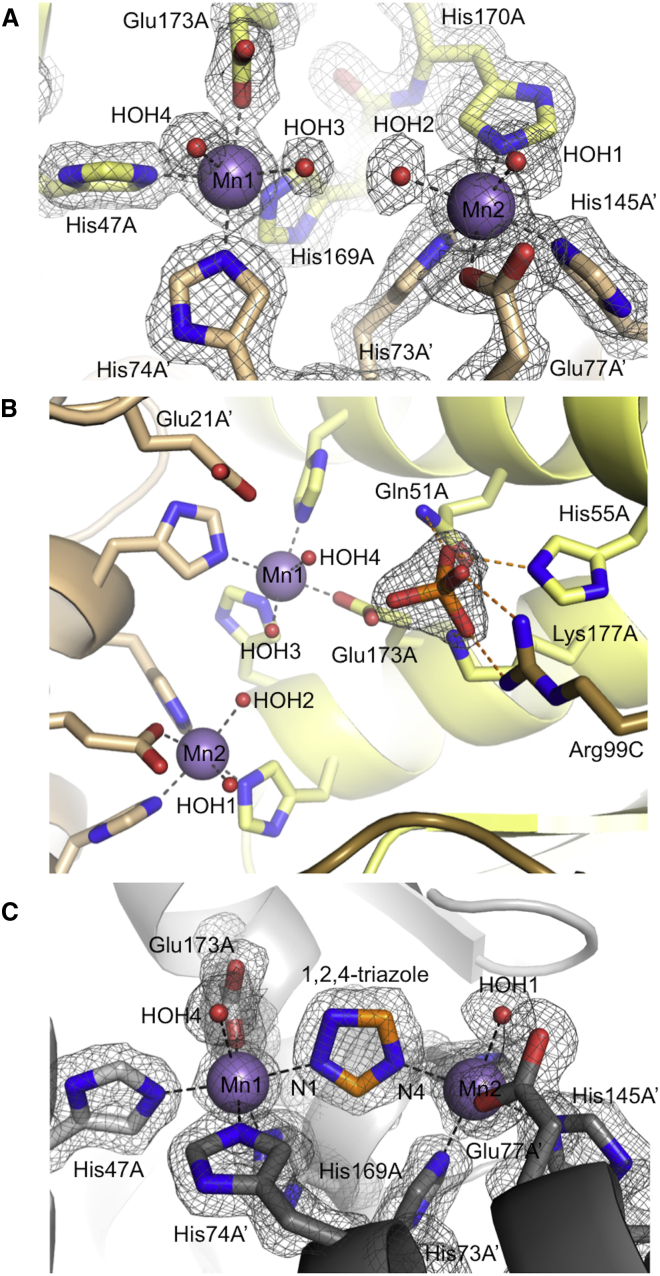
Figure 2.
Mn Coordination and Ligand Binding in the Complexes of IGPD2 with Phosphate and 1,2,4-Triazole
(A) In the holoenzyme structure, two manganese ions (Mn1 and Mn2) are octahedrally coordinated each by three histidine residues, a glutamate (from chains A and A′) and two water molecules (labeled HOH1-4). The 1.75-Å 2mFo-DFc electron density map (gray mesh) is shown surrounding the metal ions and their ligands, contoured at 1.2 σ. The protein backbone and side-chain carbon atoms are shown in yellow (chain A) or gold (chain A′), with other atoms in atom colors (N blue, O red, and P orange). The metal ions and selected water molecules are labeled and shown as purple or red spheres, respectively. Metal ion interactions are shown as black dashes.
(B) Each active site of the 24mer is constructed at the interface between three chains, labeled A, A′, and C. The phosphate binding site is surrounded by a cluster of highly conserved basic residues (Gln51A, His55A, Lys177A, and Arg99C) that are hydrogen bonded (orange dashes) to the phosphate ion. The relative position of the manganese ions and the two putative catalytic residues (Glu21A and Glu173A) are also shown, with labels for the metal ligands omitted for clarity. The atom colors, metal interactions, and electron density are shown as in (A). See also Figure S2.
(C) 1,2,4-Triazole (orange carbons) binds between the metal ions, each of which is octahedrally coordinated. The 1.3-Å 2mFo-DFc electron density map (gray mesh) surrounding the triazole is contoured at 1.5 σ. The protein backbone and side-chain carbon atoms are shown in silver with other atoms shown in atom colors as in (A). The manganese ions, coordinating waters, and metal ion interactions are labeled and shown as in (A).