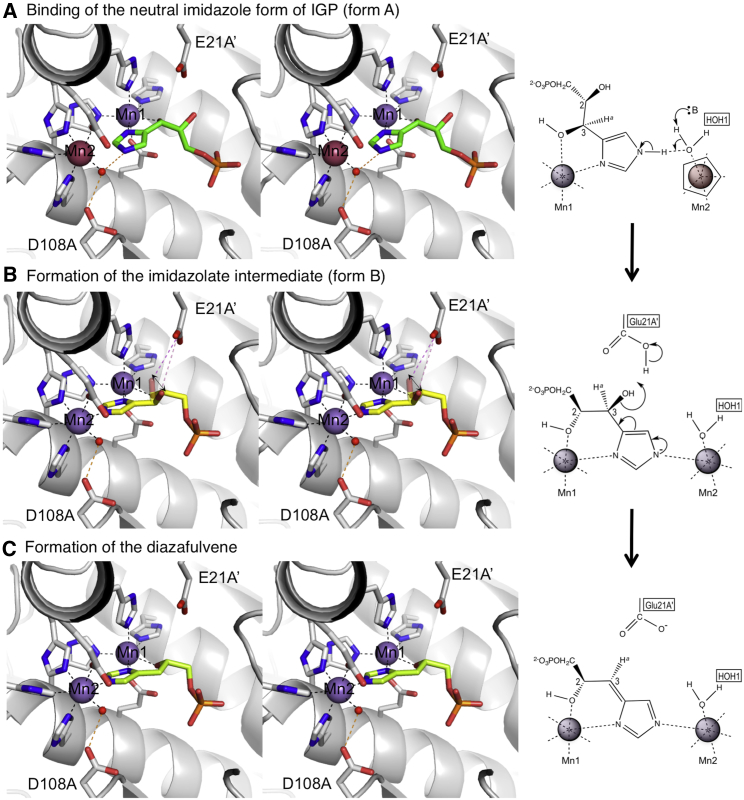
Figure 5.
The Reaction Mechanism of IGPD
(A) Stereo view to show the initial binding mode of the substrate to IGPD. IGP (green carbons) binds with a neutral imidazole ring, displacing HOH2 and leading to five-coordinate geometry around Mn2 (maroon sphere). The protonated imidazole-N3 atom forms a hydrogen bond (orange dashes) to HOH1 (red sphere), which is also hydrogen bonded to D108 and E77. The protein backbone and side-chain carbon atoms are shown in white; all non-carbon atoms are colored as in Figure 4. E21 is modeled on the basis of its position in the wild-type complexes, and other relevant side chains are shown as sticks, with labels omitted for clarity. Schematics representing the first two steps in the proposed reaction mechanism accompany the stereo views with the five-coordinate manganese ion, Mn2, colored maroon and enclosed in a pentagon. Schematic produced with ChemDraw. See also Movie S1.
(B) Ordering of the C loop (not shown) induces a conformational change, triggering deprotonation of the IGP-imidazole to imidazolate (yellow carbons) and restoring the octahedral coordination of Mn2. This is accompanied by the exchange of the C3-OH for the C2-OH as the ligand to Mn1 and repositioning of the IGP-phosphate. During substrate rearrangement between the form A and form B states, the C3-OH is transiently perpendicular (partially transparent) to the imidazolate, which is required for C3-OH elimination. A double-headed arrow represents oscillation around the intermediate. E21 is within close proximity (3.3 Å) to the C3-OH (pink dashes). Figure is drawn as in (A).
(C) The modeled position of the diazafulvene intermediate (lime green) in which the C3-OH has been eliminated. Figure is drawn as in (A).