Abstract
White-Nose Syndrome (WNS) is an epizootic disease in hibernating bats caused by the fungus Pseudogymnoascus destructans. Surveillance for P. destructans at bat hibernacula consists primarily of visual surveys of bats, collection of potentially infected bats, and submission of these bats for laboratory testing. Cryptic infections (bats that are infected but display no visual signs of fungus) could lead to the mischaracterization of the infection status of a site and the inadvertent spread of P. destructans. We determined the efficacy of visual detection of P. destructans by examining visual signs and molecular detection of P. destructans on 928 bats of six species at 27 sites during surveys conducted from January through March in 2012–2014 in the southeastern USA on the leading edge of the disease invasion. Cryptic infections were widespread with 77% of bats that tested positive by qPCR showing no visible signs of infection. The probability of exhibiting visual signs of infection increased with sampling date and pathogen load, the latter of which was substantially higher in three species (Myotis lucifugus, M. septentrionalis, and Perimyotis subflavus). In addition, M. lucifugus was more likely to show visual signs of infection than other species given the same pathogen load. Nearly all infections were cryptic in three species (Eptesicus fuscus, M. grisescens, and M. sodalis), which had much lower fungal loads. The presence of M. lucifugus or M. septentrionalis at a site increased the probability that P. destructans was visually detected on bats. Our results suggest that cryptic infections of P. destructans are common in all bat species, and visible infections rarely occur in some species. However, due to very high infection prevalence and loads in some species, we estimate that visual surveys examining at least 17 individuals of M. lucifugus and M. septentrionalis, or 29 individuals of P. subflavus are still effective to determine whether a site has bats infected with P. destructans. In addition, because the probability of visually detecting the fungus was higher later in winter, surveys should be done as close to the end of the hibernation period as possible.
Introduction
Disease surveillance in wildlife is often limited by diagnostic techniques that are cost-effective, rapid, and feasible for use on wild animals [1, 2]. For diseases where hosts display visible symptoms, visual surveys are often cost-effective and can be appealing for surveillance because they typically impose minimal disturbance on host populations [3, 4]. However, if hosts have cryptic infections that are not observable, then visual surveys will have limited utility for reliably identifying habitats harboring infected individuals (a primary goal of disease surveillance) and will underestimate infection prevalence. Estimating the efficacy of visual surveys for a particular disease is necessary to determine whether this low-cost and minimally disruptive survey method is an appropriate surveillance approach.
White-Nose Syndrome (WNS) is a rapidly spreading epizootic disease that has caused widespread declines in six species of hibernating bats in North America, raising substantial concern about the risk of extirpation and extinction of species [5–8]. WNS is caused by the fungal pathogen, Pseudogymnoascus destructans [9–11], which infects and kills bats during hibernation [12] by disrupting physiology [13–15] and natural torpor arousal patterns [10, 16]. The disease was named WNS because the faces and wings of some initially documented bats were visibly covered in white, powdery fungal growth [17]. The disease was first detected in a cave near Albany, New York in 2006, and by the spring of 2015 WNS had been confirmed in seven species of bats in 26 U.S. states and five Canadian provinces [18]. Although the exact origin of P. destructans remains unclear, recent genetic data suggest the fungus was introduced to North America from the Western Palearctic [19, 20].
Visual surveillance for WNS is conducted in hundreds of caves and mines each year and is the primary surveillance strategy recommended by the U.S. Fish and Wildlife Service WNS National Response Plan and the Canadian Wildlife Heath Cooperative WNS National Plan [21, 22]. Surveillance for WNS consists primarily of searching for bats with visible fungal infections of P. destructans (e.g. visible fungus on skin tissues), and submitting bats with suspected infection for laboratory testing by histopathology [23]. Histopathology is used to confirm the presence of epidermal cupping erosions and lesions on the wing membrane diagnostic of WNS disease [23]. Reporting of hibernacula with WNS is used to track disease spread as well as inform management decision-making, such as restricting human access to sites or requiring decontamination protocols to reduce potential spread of the fungus by humans [24].
Bats become infected with P. destructans before the fungus on skin tissues becomes visible to the human eye. These cryptic infections could easily be missed during visual surveys, causing sites to be falsely classified as ‘uninfected’ when in fact the pathogen is present and bats are infected. Falsely reporting a site as not having bats infected with P. destructans could lead to underestimates of the impact of disease on bat populations, and unrestricted human access without decontamination could lead to inadvertent spread of P. destructans. False visual detections of P. destructans caused by other fungi such as Trichophyton redellii [25] could also occur and could lead to unnecessary killing of bats for submission for histopathology. The recent development of a qPCR assay [26] to detect P. destructans DNA from epidermal swab samples from bats provides an opportunity to determine the accuracy and efficacy of visual surveys for detecting the presence of the pathogen at hibernacula and the prevalence of infection on different bat species. Although a range of different factors can affect DNA quantity extracted from swabs (e.g. extraction efficiency), this qPCR assay has been shown to be both highly specific to P. destructans and highly sensitive, making it an accurate and useful method to determine if bats are infected and for estimating prevalence [27, 28].
Our main objective was to determine the accuracy of visually detecting infections of P. destructans at bat hibernacula. Here, we define infection as the presence of P. destructans DNA detected by qPCR from swab samples collected from bats. We estimated the probability of failing to visually detect infections on bats that tested positive for P. destructans by qPCR (i.e. the probability of an infection being cryptic). We hypothesized that cryptic infections would be less likely in bats with higher pathogen loads, and as a result, cryptic infections would be more likely in species with lower pathogen loads [12]. We also compared whether the presence or absence of particular bat species at a hibernaculum increased the probability of visually detecting P. destructans on bats.
Materials and Methods
Sample collection
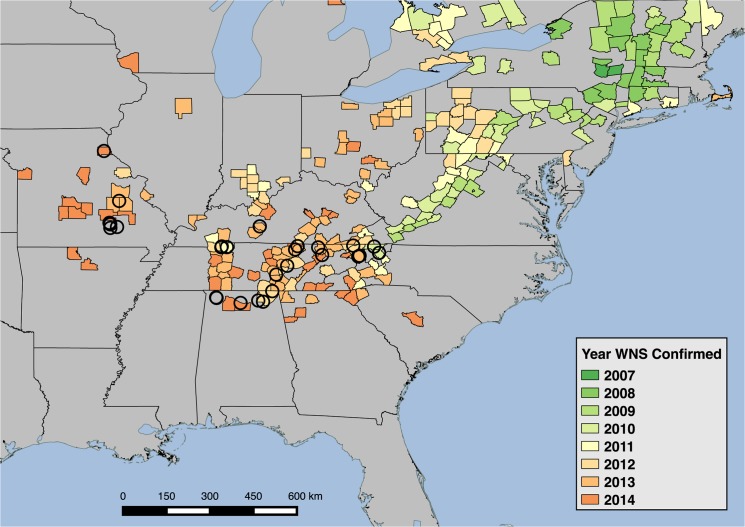
We examined the presence of P. destructans in six species in 27 hibernacula in four states in the southeastern United States (Fig 1) during winter hibernation from January through March in 2012–2014. We swabbed 928 bats of six species over three years with an average of 22 bats (range: 5–50) of one to six species present in each hibernaculum. Bats were swabbed five times on their muzzle and forearm with polyester-tipped swabs dipped in sterile water. Prior to swabbing, we noted whether fungus was visible on the bat’s skin tissues (muzzle, ears, forearms, and uropatagium) while the bat was in hand. All bats were released after sampling at the site where they had been roosting. Swabs were stored in RNAlater to preserve DNA and kept refrigerated or frozen until testing.
Fig 1. Map of sample collection.
A map of 27 hibernacula in four states where hibernating bats were sampled from January-March in 2012–2014. Shading designates the year that WNS and molecular evidence of P. destructans were confirmed in a U.S. county [18].
All bat handling procedures followed guidelines approved by the American Society of Mammalogists and the University of Tennessee Institutional Animal Care and Use Committee. Decontamination procedures issued by U.S. Fish and Wildlife Service were followed for all caving gear [24]. Permits for this research were obtained from Missouri Department of Conservation (15184, 15471, and 15871), Tennessee Wildlife Resources Agency (3716), and U.S. Fish and Wildlife Service (TE71613A-0). Other bat samples were collected in collaboration with state agency personnel with permits from Alabama Wildlife and Freshwater Fisheries and Kentucky Department of Fish and Wildlife Resources.
Sample testing
Swab samples and standards were extracted with DNeasy Blood and Tissue extraction kits (Qiagen, Valencia, CA) with modifications for fungal extractions that included the addition of lyticase during the lysis step [28]. Each extraction plate had 16 negative control wells (100% P. destructans negative) distributed throughout the plate. DNA samples were analyzed by real-time PCR using methods developed by Muller et al. [26], using a cut-off of 40 cycles for a positive detection. Cycle threshold values (Ct value) were used to calculate fungal loads, in nanograms, using the equation load = 10((22.04942-Ct value)/3.34789), which was derived from serial dilutions of a quantified standard of isolate P. destructans 20631–21. Seventy-five percent of samples were run in duplicate and a sample was considered P. destructans positive if either or both runs were positive. Fungal loads were averaged across both runs after conversion from Ct values.
Statistical analysis
Visual detection of P. destructans on bats
We used generalized linear models with a binomial distribution to determine if the probability of visually detecting P. destructans on a bat was associated with fungal load, when sampling occurred, and if detection probability differed among species. We used a bias-reduction method (package brglm in R v. 3.1.2) to deal with the complete separation present in the data (in some species no visual detections of the fungus were made). We used the number of days since January 1st to account for differences in timing of sampling as visibility of infection may increase later in the season [27, 29, 30]. We fit twelve a priori models with combinations of main, additive, and interactive effects representing our hypotheses and used Akaike information criterion (AIC) model selection criteria to determine the best-fitting model. We estimated the probability of falsely detecting visual infection using bats that tested negative by qPCR but were noted with visible white fungus in the field. We compared whether false detection differed among species using a likelihood ratio test to compare a null model to one with species included.
Visual surveys for site-level detection of P. destructans on bats
We used generalized linear models with binomial distributions in which each site visit was a data point to determine whether timing of survey, sampling effort, and species of bats examined influenced visual detection of P. destructans on bats at a site. For Myotis lucifugus, Myotis septentrionalis, and Perimyotis subflavus, we also determined whether the prevalence of infection of bats with visual infections influenced the likelihood of visually detecting the fungus during a site visit. All statistical analyses were conducted in Program R v. 3.1.2.
Results
Pathogen loads and visual detection of P. destructans on bats
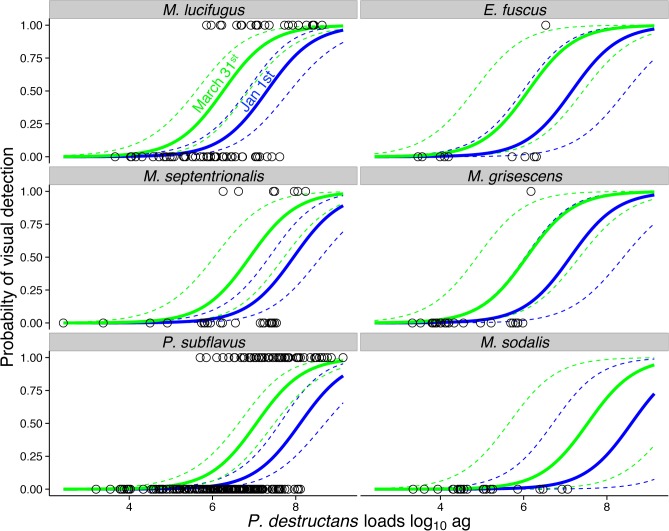
Seventy-seven percent (306/397) of bats that tested positive for P. destructans by qPCR had no visible signs of P. destructans, demonstrating that the probability of false negatives (i.e. failing to visually detect P. destructans on bats that had the pathogen) is high (Table 1). The probability of observing visible white fungus on a bat that was qPCR negative was low (14/531 or 2.6%) and did not differ among species (likelihood ratio test: χ = 5.10, df = 5; P = 0.40). The best-fitting model of the probability of visual detection included fungal load, sampling date, and an additive species effect (AIC weight = 0.55; Fitted equation for M. lucifugus = Pr(Detection) ~ -12.9 (±1.5) + 1.77 (±0.2) * log10(load) + 0.02 (±0.01) * (days since January 1); For M. septentrionalis and P. subflavus the intercept equaled -14.01 (±1.6); For the three other species (Eptesicus fuscus, Myotis grisescens, and Myotis sodalis) the intercept equaled -13.47 (±1.6)), suggesting that the probability of visually detecting P. destructans on a bat increased with pathogen load measured by qPCR, but the slope did not differ among species (Table 2, Fig 2). The probability of visually detecting P. destructans increased with the number of days since January 1st and there was only weak support that this effect differed among species (Table 2).
Table 1. Fraction of bats with visible fungus on bats tested for Pseudogymnoascus destructans by qPCR.
| Species | Fraction of bats with visible fungus | |
|---|---|---|
| qPCR + | qPCR - | |
| Eptesicus fuscus | 0.10 (1/10) | 0.0 (0/30) |
| Myotis grisescens | 0.04 (1/26) | 0.02 (5/201) |
| Myotis lucifugus | 0.35 (24/69) | 0.06 (3/50) |
| Myotis septentrionalis | 0.24 (7/29) | 0.0 (0/22) |
| Myotis sodalis | 0.0 (0/21) | 0.05 (4/76) |
| Perimyotis subflavus | 0.24 (58/242) | 0.01 (2/152) |
Sample sizes are shown in parentheses.
Table 2. Model selection results for visual detectability of Pseudogymnoascus destructans on bats.
| Model | ΔAIC | AIC weights |
|---|---|---|
| species + load + date | 0.0 | 0.55 |
| species * date + load | 2.0 | 0.20 |
| species + date * load | 2.1 | 0.19 |
| species + load | 5.2 | 0.04 |
| load | 7.1 | 0.02 |
| species * load + date | 9.7 | 0.00 |
| species * load | 18.3 | 0.00 |
| species * load * date | 29.0 | 0.00 |
| species + date | 118.8 | 0.00 |
| species | 128.6 | 0.00 |
| date | 132.4 | 0.00 |
| pd.visible.bat ~ null | 142.0 | 0.00 |
Models are ranked by ΔAIC and the best-fitting model is shown in bold.
Fig 2. Visual detectability of Pseudogymnoascus destructans on bats compared to fungal loads.
Solid lines show predicted relationships from the best-fit model (Table 2) and dashed lines show the 95% confidence bands for early (January 1st; blue lines) and late (March 31st; green lines) sampling dates. Individual circles are bats that tested positive for P. destructans by qPCR and did (y-axis value of 1) or did not (0) have visible evidence of P. destructans.
Visible infections occurred most frequently in three species (M. lucifugus, M. septentrionalis, and P. subflavus) that had the highest fungal loads and M. lucifugus had a significantly lower detectability threshold (e.g. higher intercept) compared to M. septentrionalis and P. subflavus, which were not significantly different from each other (Fig 2). Loads on the other three species (E. fuscus, M. grisescens, and M. sodalis) were usually too low to result in visible infection (Fig 2).
Efficacy of visual surveys at hibernacula
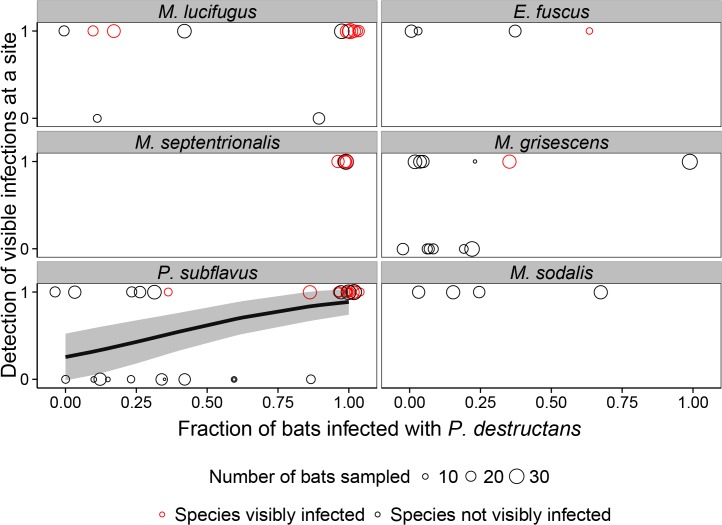
Forty percent (17/43) of sites where at least one bat tested positive for P. destructans by qPCR had no bats with visual signs of P. destructans and would have been classified as ‘uninfected’ based solely on visual surveys. The likelihood of detecting the presence of P. destructans at a site with visual surveys increased with the number of bats examined for the three species that frequently exhibit visual infections (M. lucifugus, M. septentrionalis, and P. subflavus) (Pr(Detection) ~ -0.90 + 0.12 (± 0.051) * #mylu.myse.pesu.sampled; N = 43; P = 0.02), and there was very weak support for the influence by when a visit occurred between January and March or examination of other species (Table 3). The probability of visual detection of P. destructans at a site increased with prevalence of infection for P. subflavus (Pr(Detection) ~ -1.1 + 3.13 (± 0.051) * Prevalence; N = 32; P < 0.01), but not for M. lucifugus (Pr(Detection) ~ -1.1 + 1.0 (± 1.8) * Prevalence; N = 13; P = 0.56) or M. septentrionalis, the latter of which had a prevalence of 100% at all sites (Fig 3). Visual surveys that include either 17 M. lucifugus or 17 M. septentrionalis have a 99% likelihood of detecting P. destructans if it is present at the site. For P. subflavus, examining at least 29 bats is required to have a 99% chance of detecting P. destructans if it is present.
Table 3. Model selection results for visual detectability of Pseudogymnoascus destructans at hibernacula.
#mylu.myse.pesu.sampled refers to the sum of the number of bats of three species sampled (M. lucifugus – mylu, M. septentrionalis – myse, P. subflavus – pesu).
| Model | ΔAIC | AIC weights |
|---|---|---|
| #mylu.myse.pesu.sampled | 0.0 | 0.56 |
| date + #mylu.myse.pesu.sampled | 1.3 | 0.29 |
| null | 4.9 | 0.05 |
| all.bats.sampled | 5.0 | 0.05 |
| all.bats.sampled + date | 5.6 | 0.03 |
| date | 6.6 | 0.02 |
Models are ranked by ΔAIC and the best-fitting model is shown in bold.
Fig 3. Detection of visible Pseudogymnoascus destructans on bats at hibernacula and the fraction of bats with P. destructans at that site as determined by qPCR.
Circles represent sites where a species was sampled, with red circles indicating sites where at least one individual of that species had visible fungus and black circles indicating sites where no individuals of that species were observed with visible fungus. The size of the circles is scaled to the number of bats sampled at a site. The x-axis shows the proportion of bats that were positive for P. destructans by qPCR and the y-axis shows whether at least one individual bat at that site of any species was negative (0) or positive (1) for visible fungal infections. Prevalence of infection was a significant predictor for detection of visible infections at a site for a single species, P. subflavus. Solid black line and gray shading for P. subflavus represent the best-fit line and 95% confidence band for the relationship between prevalence of infection and detection of visible infections at hibernacula.
Discussion
Our results suggest that cryptic infections are widespread and that solely using visible signs of P. destructans greatly underestimates infection prevalence in bats even during mid to late winter (January-March) when the majority of surveillance surveys for WNS are conducted. Cryptic infections were so common in some species (E. fuscus, M. grisescens, and M. sodalis) that visual surveys were only useful for detecting P. destructans at a site if other species (M. lucifugus, M. septentrionalis, and/or P. subflavus) also were present and examined. The higher percentages of the latter three species that displayed visible P. destructans, combined with the high infection prevalence in these species, resulted in a very high likelihood that P. destructans was detected at a site whenever these bat species were present.
Our results also show that the probability of visual detection increases with fungal load of P. destructans, and differences in fungal loads among species explain most of the differences in the probability of observing visible P. destructans on bats. This is likely because higher loads indicate a larger number of conidia and hyphae on the bats and a greater likelihood of the fungus being visible. This is consistent with the finding that the probability of visual detection of P. destructans on bats was higher later in the hibernation season when the fungus has had sufficient time to grow on the bats and is at maximal loads [12], suggesting that visual surveys should be scheduled late in hibernation to be maximally effective. Our findings are similar to patterns of visual prevalence in Europe where visible infections also peaked in late hibernation [29]. Hibernation season length may influence visual detection given that most bats become infected at the start of hibernation and fungal loads increase once bats are torpid [12]. Thus, infections may become visible sooner in northern latitudes where bats likely enter hibernation earlier [31].
Even with the same fungal load, some species were more likely to exhibit visible P. destructans (Fig 2). Visible P. destructans was detected at significantly lower loads on M. lucifugus than other species (Fig 2), perhaps because their darker skin provides more visual contrast with the white fungus. Myotis lucifugus and M. septentrionalis, when present, are the best ‘sentinels’ or indicators of the presence of P. destructans when surveying for visible signs, and surveying P. subflavus can also be useful. In contrast, fungal loads in E. fuscus, M. grisescens, and M. sodalis are simply too low to consistently result in visible P. destructans. Differences in fungal loads and infection intensity among species suggests interesting differences in either transmission, hibernating behaviors, and/or disease susceptibility among hibernating species exposed to P. destructans [5, 32, 33].
Currently, visual surveys are routinely used to determine whether P. destructans has invaded new hibernacula [21, 22]. Our results show that the efficacy of these visual surveys depends on which species are present at a site and how many bats are examined for visible fungus. For example, the presence of M. lucifugus or M. septentrionalis increases the probability that P. destructans can be detected visually at a site and that these can be used as ‘sentinel’ species for the presence of P. destructans (Fig 3). Our results suggest that with a moderate survey effort of examining either 20 (if surveying M. lucifugus or M. septentrionalis) or 30 (if only P. subflavus are examined) individuals at a site, then visual surveys can indeed be effective at determining whether bats are infected with P. destructans at a hibernaculum. At sites with species that rarely or never have visible signs of P. destructans, such as E. fuscus, M. grisescens, or M. sodalis, visual surveys are ineffective. To ensure visual surveillance is effective at determining whether P. destructans has invaded new sites [21, 22], future surveillance guidelines should incorporate these specific recommendations on species and sample sizes required for effective surveillance efforts.
The widespread occurrence of cryptic infections in all species has direct relevance to management and surveillance of this disease [34, 35]. Visual surveys can be an effective and relatively low-cost part of surveillance activities, especially in areas where routine winter colony counts are already conducted [36], only as long as sites contain sufficient numbers of bats (>20) of species that exhibit visual infections (e.g. M. lucifugus, M. septentrionalis, and/or P. subflavus). Further, visual surveys of individual bats are most effective late in the hibernation season. However, for detection of P. destructans on species with predominately cryptic infections and to accurately measure prevalence, swab sampling and testing samples with molecular methods are needed [12, 26, 27]. Ultraviolet (UV) illumination has recently been proposed for WNS surveillance based on comparisons with histological examination of bats submitted for testing based on visual signs of WNS and bats collected in areas where the fungus has been present for several years [37]. We did not examine bats under UV illumination and a comparison of this method with molecular testing of swab samples would be useful to determine whether UV illumination is effective for detecting cryptic infections on the leading edge of fungal invasion. Currently, the U.S. Fish and Wildlife Service WNS National Response Plan and the Canadian Wildlife Health Cooperative WNS National Plan surveillance protocols rely entirely on visual surveillance [21, 22], but our findings suggest that combining swab sampling and visual surveys would improve national surveillance of this disease.
There are currently no active management strategies for control or mitigation of WNS other than cave closures [21, 34]. However, activities such as culling have been considered as a means to prevent the spread of the disease to new regions [38]. The occurrence of cryptic infections demonstrates that culling visibly infected bats will be ineffective at halting the spread of P. destructans, supporting early modeling efforts [38]. Further, recent evidence suggests that culling infected individuals, even using a highly sensitive method (e.g. qPCR on swab samples), will be ineffective because P. destructans is often widespread in the environment a year after the fungus reaches a site, and can persist at sites and in the absence of bats for long periods [27, 38–41]. Our findings that cryptic infections commonly occur at bat hibernacula suggest that although the spread of P. destructans across North America is consistent with spread by bats [42–44], restricting recreational access and requiring field hygiene protocols to decontaminate gear will reduce potential human-mediated spread.
Acknowledgments
We thank 3 anonymous reviewers for insightful feedback on earlier drafts and personnel from the following agencies/organizations for assistance in the field and access to hibernacula: U.S. Fish and Wildlife Service, U.S. National Park Service, Tennessee Wildlife Resources Agency, Tennessee Valley Authority, Missouri Department of Conservation, Alabama Wildlife and Freshwater Fisheries, Kentucky Department of Fish and Wildlife Resources, Southeastern Cave Conservancy, the Nature Conservancy, the Missouri Bat Census, and Environmental Solutions and Innovations. We thank Darwin Brack for assistance with swab sample collection. We thank Kevin Dress and Nicolette Janke for lab work on DNA extractions and qPCR. We thank David Blehert and Jeff Lorch for providing P. destructans samples for creating standards.
Data Availability
All relevant data are available from Dryad Digital Repository (doi:10.5061/dryad.8mm58).
Funding Statement
Funding was provided by the National Science Foundation Ecology of Infectious Diseases (DEB-1115895 and DEB-1336290) to WFF, AMK, JTF, and GFM; and the University of Tennessee Department of Ecology and Evolutionary Biology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stallknecht DE. Impediments to wildlife disease surveillance, research, and diagnostics In: Childs JE, Mackenzie JS, Richt JA, editors. Wildlife and emerging zoonotic diseases: the biology, circumstances and consequences of cross-species transmission. Berlin: Springer; 2007. pp. 445–461. [DOI] [PubMed] [Google Scholar]
- 2. Sleeman JM. Has the time come for big science in wildlife health? Ecohealth. 2013; 10:335–338. 10.1007/s10393-013-0880-0 [DOI] [PubMed] [Google Scholar]
- 3. Hochachka WM, Dhondt AA. Density-dependent decline of host abundance resulting from a new infectious disease. Proc Natl Acad Sci USA. 2000; 97:5303–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blanchong JA, Samuel MD, Goldberg DR, Shadduck DJ, Lehr MA. Persistence of Pasteurella multocida in wetlands following avian cholera outbreaks. J Wildl Dis. 2006; 42:33–39. [DOI] [PubMed] [Google Scholar]
- 5. Langwig KE, Frick WF, Bried JT, Hicks AC, Kunz TH, Kilpatrick AM. Sociality, density-dependence and microclimates determine the persistence of populations suffering from a novel fungal disease, white-nose syndrome. Ecol Lett. 2012; 15:1050–1057. 10.1111/j.1461-0248.2012.01829.x [DOI] [PubMed] [Google Scholar]
- 6. Frick WF, Puechmaille SJ, Hoyt JR, Nickel BA, Langwig KE, Foster JT, et al. Disease alters macroecological patterns of North American bats. Global Ecol Biogeogr. 2015; 10.1111/geb.12290 [DOI] [Google Scholar]
- 7. Frick WF, Pollock JF, Hicks AC, Langwig KE, Reynolds DS, Turner GG, et al. An emerging disease causes regional population collapse of a common North American bat species. Science. 2010; 329:679–682. 10.1126/science.1188594 [DOI] [PubMed] [Google Scholar]
- 8. Thogmartin WE, Sanders-Reed CA, Szymanski JA, McKann PC, Pruitt L, King RA, et al. White-nose syndrome is likely to extirpate the endangered Indiana bat over large parts of its range. Biol Conserv. 2013; 160:162–172. [Google Scholar]
- 9. Lorch JM, Meteyer CU, Behr MJ, Boyles JG, Cryan PM, Hicks AC, et al. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature. 2011; 480:376–379. 10.1038/nature10590 [DOI] [PubMed] [Google Scholar]
- 10. Warnecke L, Turner JM, Bollinger TK, Lorch JM, Misra V, Cryan PM, et al. Inoculation of bats with European Geomyces destructans supports the novel pathogen hypothesis for the origin of white-nose syndrome. Proc Natl Acad Sci USA. 2012; 109:6999–7003. 10.1073/pnas.1200374109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Minnis AM, Linder DL. Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biol. 2013; 117:638–649. 10.1016/j.funbio.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 12. Langwig KE, Frick WF, Reynolds R, Parise KL, Drees KP, Hoyt JR, et al. Host and pathogen ecology drive the seasonal dynamics of a fungal disease, white-nose syndrome. Proc R Soc B Biol Sci. 2015; 282:20142335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Warnecke L, Turner JM, Bollinger TK, Misra V, Cryan PM, Blehert DS, et al. Pathophysiology of white-nose syndrome in bats: a mechanistic model linking wing damage to mortality. Biol Lett. 2013; 9:20130177 10.1098/rsbl.2013.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cryan PM, Meteyer CU, Blehert DS, Lorch JM, Reeder DM, Turner GG, et al. Electrolyte depletion in white-nose syndrome bats. J Wildl Dis. 2013; 49:398–402. 10.7589/2012-04-121 [DOI] [PubMed] [Google Scholar]
- 15. Verant ML, Meteyer CU, Speakman JR, Cryan PM, Lorch JM, Blehert DS. White-nose syndrome initiates a cascade of physiologic disturbances in the hibernating bat host. BMC Physiol. 2014; 14:10 10.1186/s12899-014-0010-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reeder DM, Frank CL, Turner GG, Meteyer CU. Frequent arousal from hibernation linked to severity of infection and mortality in bats with White-Nose Syndrome. PLoS One. 2012; 7:e38920 10.1371/journal.pone.0038920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blehert DS, Hicks AC, Behr M, Meteyer CU, Berlowski-Zier BM, Buckles EL, et al. Bat White-Nose Syndrome: An emerging fungal pathogen? Science. 2009; 323:227 10.1126/science.1163874 [DOI] [PubMed] [Google Scholar]
- 18.U.S. Fish and Wildlife Service. White-nose syndrome map. 2015. Available: http://www.whitenosesyndrome.org/resources/map
- 19. Puechmaille SJ, Frick WF, Kunz TH, Racey PA, Voigt CC, Wibbelt G, et al. White-nose syndrome: is this emerging disease a threat to European bats? Trends Ecol Evol. 2011; 26:570–576. 10.1016/j.tree.2011.06.013 [DOI] [PubMed] [Google Scholar]
- 20. Leopardi S, Blake D, Puechmaille SJ. White-Nose Syndrome fungus introduced from Europe to North America. Curr Biol. 2015; 25:R217–R219. 10.1016/j.cub.2015.01.047 [DOI] [PubMed] [Google Scholar]
- 21.U.S. Fish and Wildlife Service. A national plan for assisting states, federal agencies, and tribes in managing White-Nose Syndrome in bats. 2011. Available: https://www.whitenosesyndrome.org/sites/default/files/white-nose_syndrome_national_plan_may_2011.pdf
- 22.Canadian Wildlife Health Cooperative. A national plan to manage White Nose Syndrome in bats in Canada. 2015. Available: http://www.cwhc-rcsf.ca/docs/BatWhiteNoseSyndrome-NationalPlan.pdf.
- 23. Meteyer CU, Buckles EL, Blehert DS, Hicks AC, Green DE, Shearn-Bochsler V, et al. Histopathologic criteria to confirm white-nose syndrome in bats. J Vet Diagn Invest. 2009; 21:411–414. [DOI] [PubMed] [Google Scholar]
- 24. Shelley V, Kaiser S, Shelley E, Williams T, Kramer M, Haman K, et al. Evaluation of strategies for the decontamination of equipment for Geomyces destructans, the causative agent of White-Nose Syndrome (WNS). J Cave Karst Stud. 2013; 75:1–10. [Google Scholar]
- 25. Lorch JM, Minnis AM, Meteyer CU, Redell JA, White JP, Kaarakka HM, et al. The fungus Trichophyton redellii sp. nov. causes skin infections that resemble White-Nose Syndrome of hibernating bats. J Wildl Dis. 2015; 51:36–47. 10.7589/2014-05-134 [DOI] [PubMed] [Google Scholar]
- 26. Muller LK, Lorch JM, O’Connor M, Gargas A, Blehert DS. Bat white-nose syndrome: a real-time TaqMan polymerase chain reaction test targeting the intergenic spacer region of Geomyces destructans . Mycologia. 2013; 105:253–259. 10.3852/12-242 [DOI] [PubMed] [Google Scholar]
- 27. Langwig KE, Hoyt JR, Parise KL, Kath J, Kirk D, Frick WF, et al. Disease dynamics of white-nose syndrome invasion, Midwest USA. Emerg Infect Dis. 2015; 21:1023–1026. 10.3201/eid2106.150123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shuey MM, Drees K, Lindner DL, Keim P, Foster JT. A highly sensitive qPCR assay for the detection and differentiation of Pseudogymnoascus destructans and Pseudogymnoascus species. Appl Environ Microbiol. 2014; 80:1726–1731. 10.1128/AEM.02897-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Puechmaille SJ, Wibbelt G, Korn V, Fuller H, Forget F, Mühldorfer K, et al. Pan-European distribution of white-nose syndrome fungus (Geomyces destructans) not associated with mass mortality. PLoS One. 2011; 10.1371/journal.pone.0019167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sachanowicz K, Stępień A, Ciechanowski M. Prevalence and phenology of white-nose syndrome fungus Pseudogymnoascus destructans in bats from Poland. Cent Eur J Biol. 2014; 10.2478/s11535-013-0280-z [DOI] [Google Scholar]
- 31. Norquay KJO, Willis CKR. Hibernation phenology of Myotis lucifugus . J Zool. 2014; 294:85–92. [Google Scholar]
- 32. Frank CL, Michalski A, McDonough AA, Rahimian M, Rudd RJ, Herzog C. The resistance of a North American bat species (Eptesicus fuscus) to White-Nose Syndrome. PLoS One. 2014; 10.1371/journal.pone.0113958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson JS, Reeder DM, Lilley TM, Czirják GÁ, Voigt CC, McMichael JW, et al. Antibodies to Pseudogymnoascus destructans are not sufficient for protection against white-nose syndrome. Ecol Evol. 2015; 5:2203–2214. 10.1002/ece3.1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langwig KE, Voyles J, Wilber MQ, Frick WF, Murray KA, Bolker BM, et al. Context-dependent conservation responses to emerging wildlife diseases. Front Ecol Environ. 2015; 13:195–202. [Google Scholar]
- 35. Foley J, Clifford D, Castle K, Cryan PM, Ostfeld RS. Investigating and managing the rapid emergence of white nose syndrome, a novel, fatal, infectious disease of hibernating bats. Conserv Biol. 2011; 25:223–231. 10.1111/j.1523-1739.2010.01638.x [DOI] [PubMed] [Google Scholar]
- 36. Loeb SC, Rodhouse TJ, Ellison LE, Lausen CL, Reichard JD, Irvine KM, et al. A plan for the North American bat monitoring program (NABat). USDA Forest Service Research and Development Southern Research Station. 2015. Available: http://www.srs.fs.usda.gov/pubs/gtr/gtr_srs208.pdf. [Google Scholar]
- 37. Turner GG, Meteyer CU, Barton H, Gumbs JF, Reeder DM, Overton B, et al. Nonlethal screening of bat-wing skin with the use of ultraviolet fluorescence to detect lesions indicative of white-nose syndrome. J Wildl Dis. 2014; 50:566–573. 10.7589/2014-03-058 [DOI] [PubMed] [Google Scholar]
- 38. Hallam TG, McCracken GF. Management of the panzootic White-Nose Syndrome through culling of bats. Conserv Biol. 2011; 25:189–194. 10.1111/j.1523-1739.2010.01603.x [DOI] [PubMed] [Google Scholar]
- 39. Lorch JM, Muller LK, Russell RE, O'Connor M, Lindner DL, Blehert DS. Distribution and environmental persistence of the causative agent of white-nose syndrome, Geomyces destructans, in bat hibernacula of the Eastern United States. Appl Environ Microbiol. 2013; 79:1293–1301. 10.1128/AEM.02939-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoyt JR, Okoniewski J, Langwig KE, Frick WF, Stone WB, Kilpatrick AM. Long-term persistence of Pseudogymnoascus destructans, the causative agent of white-nose syndrome, in the absence of bats. Ecohealth. 2014; 10.1007/s10393-014-0981-4 [DOI] [PubMed] [Google Scholar]
- 41. Reynolds HT, Ingersoll T, Barton HA. Modeling the environmental growth of Pseudogymnoascus destructans and its impact on the white-nose syndrome epidemic. J Wildl Dis. 2015; 51:318–331. 10.7589/2014-06-157 [DOI] [PubMed] [Google Scholar]
- 42. Wilder AP, Frick WF, Langwig KE, Kunz TH. Risk factors associated with mortality from white-nose syndrome among hibernating bat colonies. Biol Lett. 2011; 7:950–953. 10.1098/rsbl.2011.0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maher SP, Kramer AM, Pulliam JT, Zokan MA, Bowden SE, Barton HD, et al. Spread of white-nose syndrome on a network regulated by geography and climate. Nat Commun 2012; 3:1306–1308. 10.1038/ncomms2301 [DOI] [PubMed] [Google Scholar]
- 44. O’Regan SM, Magori K, Pulliam JT, Zokan MA, Kaul RRB, Barton HD, et al. Multi-scale model of epidemic fadeout: will local extirpation events inhibit the spread of white-nose syndrome? Ecol Appl. 2015; 25:621–633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are available from Dryad Digital Repository (doi:10.5061/dryad.8mm58).