Figure 3.
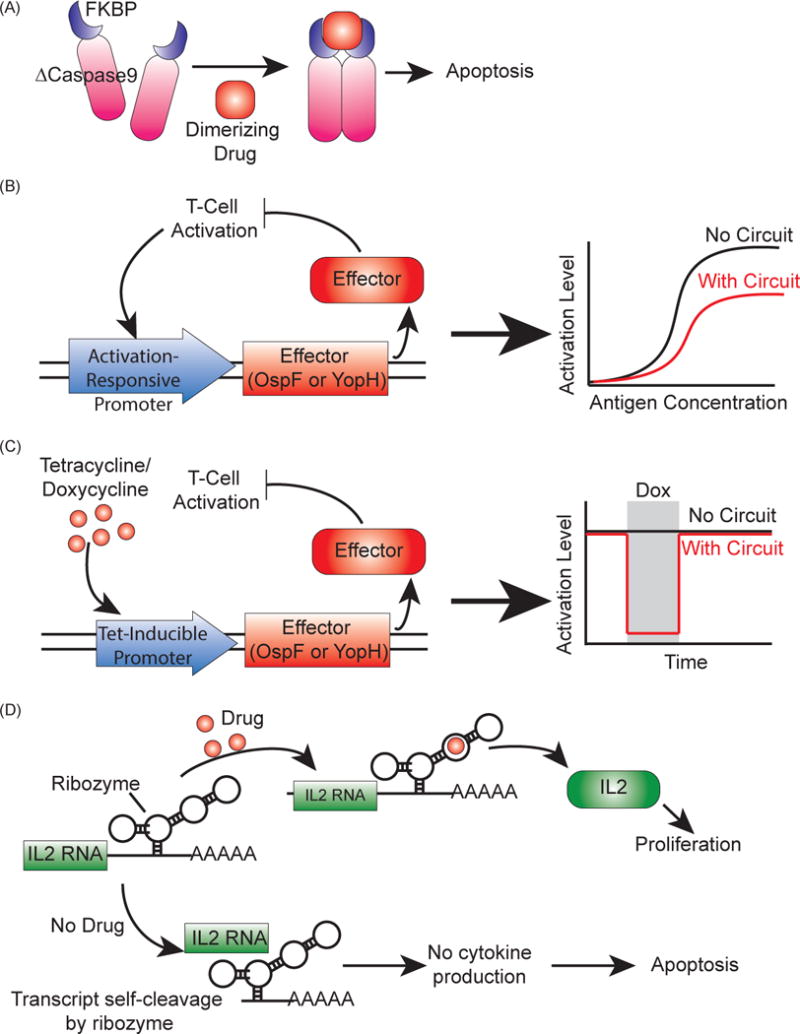
Synthetic genetic circuits to regulate T cell activity in patients. (A) Inducible suicide gene using iCasp9, a Caspase9 mutant (ΔCaspase9) fused to FKBP dimerizing domains [63, 66]. When the dimerizing drug AP1903 is added, ΔCaspase9 dimerizes and drives apoptosis. (B) Amplitude limiter using the bacterial virulence proteins OspF or YopH as effectors [70]. These effectors reduce T cell activation, and expressing them under an activation-responsive promoter creates a negative feedback loop that reduces T cell activation. (C) Pause switches using OspF or YopH as effectors [70]. Using a tetracycline-inducible promoter to control effector expression, the addition of the drug will drive effector production, which will in turn shut off activation until the drug is removed. (D) A ribozyme switch to control T-cell proliferation [71]. Cytokine RNA is expressed with the ribozyme switch, which will drive self-cleavage of the transcript and lead to no cytokine expression without the addition of the appropriate drug. When drug is added, the cytokine transcript is preserved, leading to cytokine production and proliferation.
