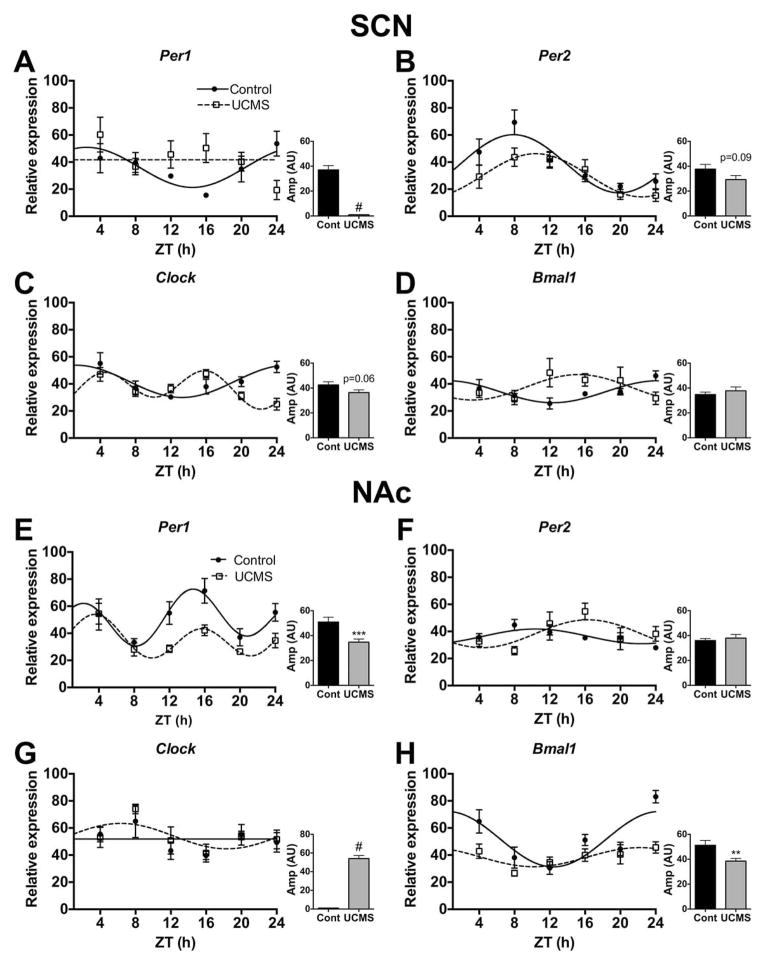
Figure 6.
UCMS alters diurnal rhythms of circadian gene expression in the SCN and NAc of mice. Brains were collected across the day (ZT4-24) and the SCN and NAc were micropunched for gene expression analyses. Chronic stress altered the diurnal rhythms of gene expression for the core circadian genes, including Per1, Per2, Clock, and Bmal1 in the SCN (A–D), and in the NAc (E–H). Sine wave fits using linear harmonic regression with an assumed period of 24 h for control and UCMS mice (solid and dotted lines, respectively) are superimposed with group means ± SEM for each ZT. Straight line denotes no significant curve fit, and thus a lack of detectable rhythms. All curve fits are significant (p<0.05). **, p<0.01 and ***, p<0.001, unpaired t-tests between control and UCMS amplitude (graph inlets). #, represents no amplitude value due to an undetectable rhythm.