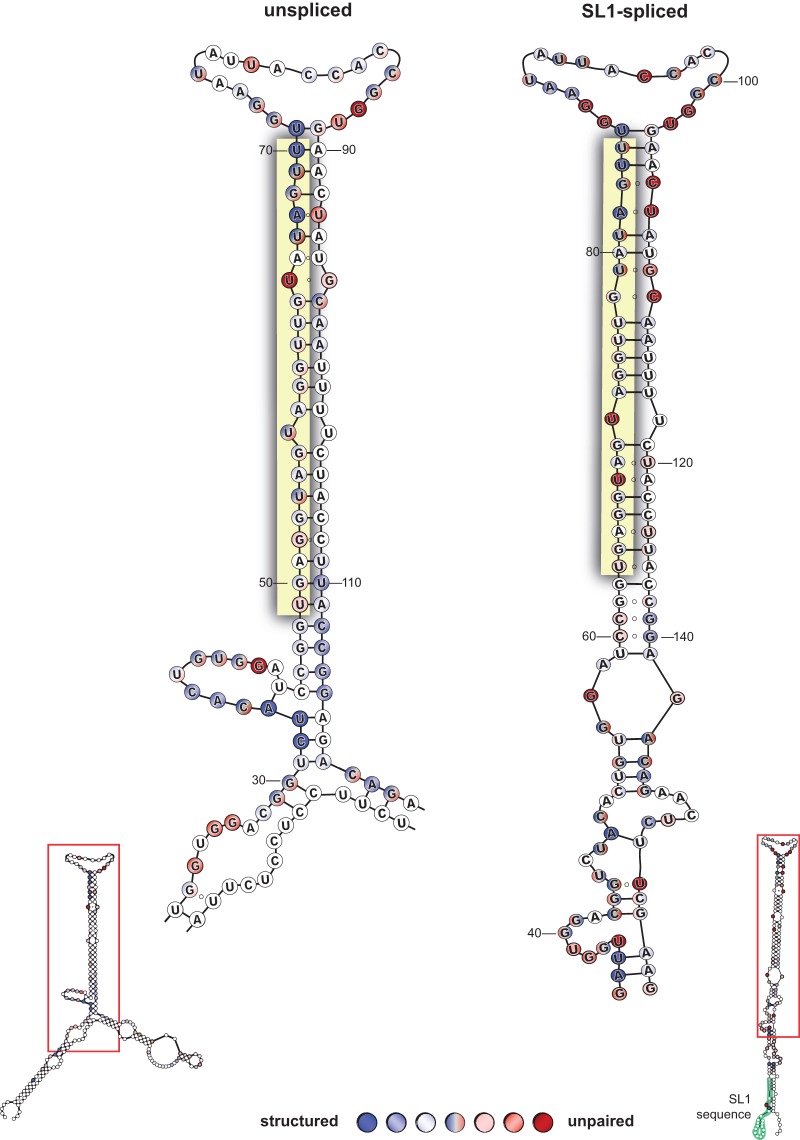
FIGURE 1.
Secondary structures of unspliced and spliced pri-let-7 RNAs. RNA secondary structure analysis of in vitro transcribed unspliced and SL1-spliced model pri-let-7 transcripts. Numbers indicate positions of bases in the model transcripts. The composite results from four independent experiments are shown. Here we show a close-up view of the area in the red box subject to processing by the Microprocessor. Three shades of color are used to represent “high,” “medium,” and “low” sensitivity of each nucleotide to the RNase enzyme, such that the darker the color the more reactive to cleavage and the more “structured” (blue) or “unpaired” (red) the base appears to be. Nucleotides in shades of blue circles were sensitive to cleavage by RNase V1, denoting double-strandedness. Shades of red circles represent single-stranded nucleotides that were cleaved in response to RNase T1 or RNase A treatment. Nucleotides labeled both red and blue indicate evidence for single- and double-stranded nature, which is expected from G–U pairs and regions of the transcript that are more dynamic. Unlabeled (white) nucleotides were inconclusive. Open circles between nucleotides represent regions of expected pairing that were not detectable by the RNase structure probing. The mature let-7 miRNA sequence is highlighted in yellow. The SL1 sequence is highlighted in green and labeled. Representative PAGE used for data analyses are shown in Supplemental Figure 1.