FIGURE 3.
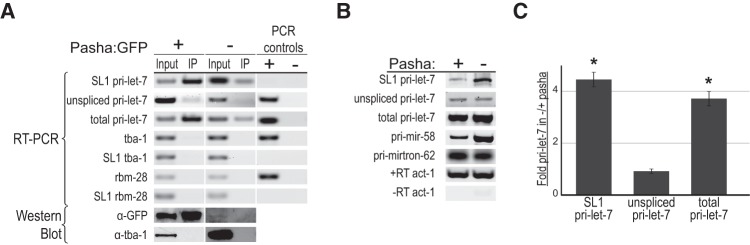
The Microprocessor prefers SL1-spliced pri-let-7 for processing in vivo. (A) Detection of transcripts associated with the Microprocessor in vivo. Extracts from L4 stage worm strains with (+) and without (−) the rescuing Pasha:GFP transgene were subjected to Immunoprecipitation (IP) using an anti-GFP antibody. Total (Input) and Immunoprecipitated (IP) RNAs were detected by RT-PCR followed by Agarose gel electrophoresis. PCR controls show the results using genomic DNA (+), which should not detect SL1-spliced sequences, or water (−) as templates. The bottom panels show the results of Western blot analysis with a GFP antibody to monitor the IP of Pasha:GFP and tubulin (TBA-1) as a loading and specificity control. Results are representative of two independent IP experiments. (B) Total RNA from pasha mutant (−) and rescued (+) worms was collected from L4 staged animals and used for RT-PCR analysis of the indicated transcripts. Results were analyzed by agarose gel electrophoresis and represent three independent experiments. Actin (act-1) served both as a loading control and to control for genomic DNA contamination in –RT samples. Pri-miR-58 is a constitutively expressed miRNA subject to Pasha regulation. Pri-mirtron-62 is not part of the canonical miRNA biogenesis pathway. (C) Average fold increase of pri-let-7 isoforms in pasha mutants (−) relative to rescued (+) worms collected at L4 and analyzed by qRT-PCR. Each pri-let-7 isoform was first normalized to the control transcript Y45F10D.4. The error bars represent standard error. (*) P < 0.05.
