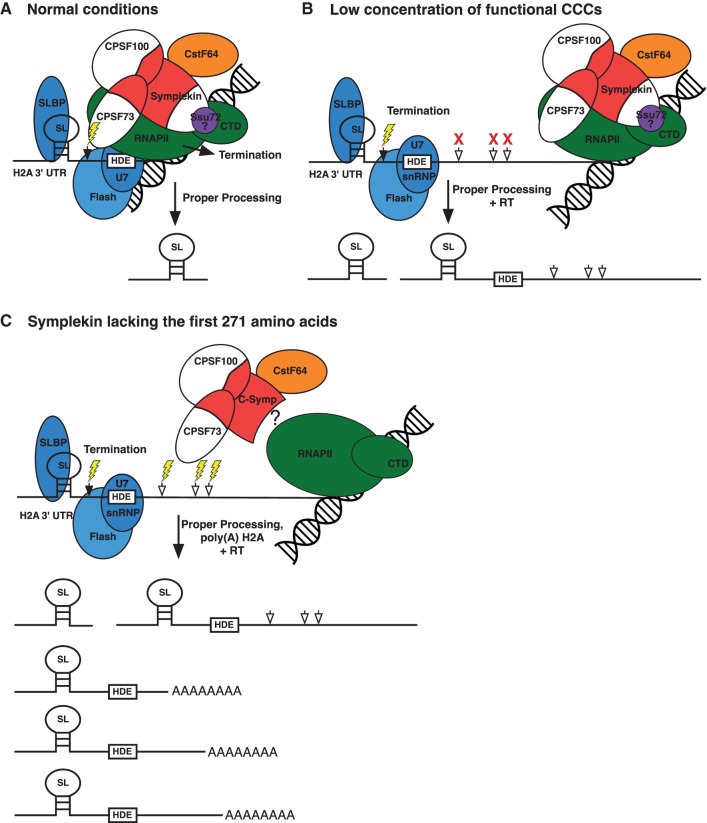
FIGURE 6.
Histone mRNA 3′ end processing models. (A) A model for cotranscriptional histone mRNA 3′ end processing under normal conditions is shown. Following transcription of the cleavage site (black arrow) and HDE, RNAPII pauses allowing recruitment and proper orientation of the CCC, SLBP, U7 snRNP, and FLASH to the cleavage site. This results in H2A mRNA processing at the most proximal cleavage site (lightning bolt) and transcription termination. (B) A model for cotranscriptional histone mRNA 3′ end processing under low CCC concentration conditions is shown. If a CCC can be cotranscriptionally recruited to the cleavage site (black arrow), the H2A mRNA 3′ end will be properly processed (lightning bolt). When no functional CCC can be recruited, RNAPII continues transcribing and use of downstream poly(A) sites (white arrows) is inefficient resulting in read through (RT) H2A mRNAs. (C) A model for H2A mRNA 3′ end processing by CCCs containing Symplekin lacking the first 271 amino acids is shown. A high concentration of functional CCCs containing N-terminally truncated Symplekin can use downstream poly(A) sites (white arrows) for H2A mRNA 3′ end processing. We hypothesize that the first 271 amino acids of Symplekin mediate cotranscriptional histone mRNA 3′ end formation.